ENCAB000AXU
Antibody against Homo sapiens CTCF
Homo sapiens
K562, liver
characterized to standards
Homo sapiens
SK-N-SH, HepG2, neural cell, sigmoid colon, transverse colon
characterized to standards with exemption
Homo sapiens
any cell type or tissue
partially characterized
- Status
- released
- Source (vendor)
- Millipore
- Product ID
- 07-729
- Lot ID
- DAM1745366
- Characterized targets
- CTCF (Homo sapiens)
- Host
- rabbit
- Clonality
- polyclonal
- Purification
- crude
- Antigen description
- Raised against KLH-conjugated, synthetic peptide corresponding to amino acids 659-675 ( C-TNQPKQNQPTAIIQVED) of human CCCTC-binding factor (CTCF) with a N-terminal cysteine added for cunjugation purposes
- Antigen sequence
- C-TNQPKQNQPTAIIQVED
- External resources
Characterizations
CTCF (Homo sapiens)
Method: immunoprecipitation followed by mass spectrometry
compliant
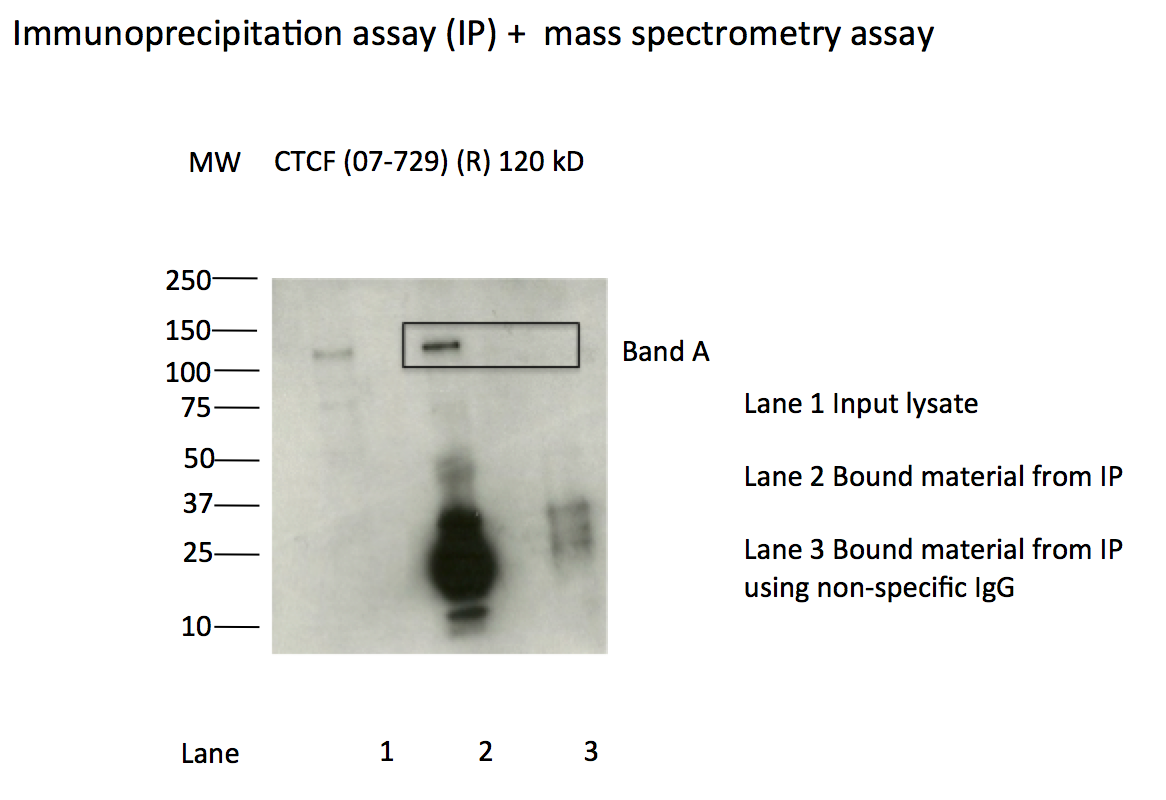
- Caption
- IP followed by mass spectrometry: Briefly, protein was immunoprecipitated from K562 whole cell lysates using 07-729, and the IP fraction was loaded on a 10% polyacrylamide gel (NuPAGE Bis-Tris Gel) and separated with an Invitrogen NuPAGE electrophoresis system. The gel was silver-stained, gel fragments corresponding to the bands indicated were excised and destained using the SilverSNAP Stain for Mass Spectrometry (Pierce). Then proteins were treated with trypsin using the in-gel digestion method. Digested proteins were analyzed on a LTQ-Orbitrap (Thermo Scientific) b the nanoLC-ESI-MS/MS technique. Peptides were identified by the SEQUEST algorithm and filtered with a high confidence threshold (protein false discovery rate <1%, 2 peptides per protein minimum).
- Submitted by
- Kathrina Onate
- Lab
- Michael Snyder, Stanford
- Grant
- U54HG004558
- Download
- CTCF_final_AXU.pdf
CTCF (Homo sapiens)
K562
compliant
- Caption
- Immunoprecipitation of CTCF from K562 cells using 07-729. Lane 1: input nuclear lysate. Lane 2: material immunoprecipitated with 07-729. Lane 3: material immunoprecipitated using control IgG. Band A was excised from gel and subject to analysis by mass spectrometry. Expected band size ~120 kD.
- Submitted by
- Kathrina Onate
- Lab
- Michael Snyder, Stanford
- Grant
- U54HG004558
- Download
- CTCF_IP.png
CTCF (Homo sapiens)
liver
compliant
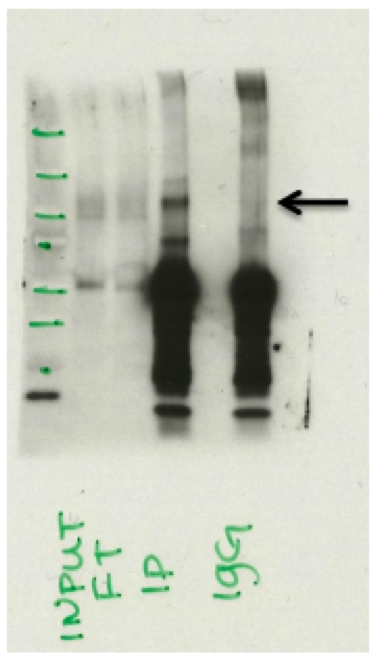
- Caption
- Western Blot analysis of immunoprecipitated proteins from Human Liver nuclear extracts detected using anti-CTCF (07-729) antibody from Millipore. Expected protein band 120kD. Input: Input lysate, FT: Flowthrough from IP, IP: Immunoprecipitated fraction from 07-729, IgG: Control Immunoprecipitation using IgG.
- Submitted by
- Esther Chan
- Lab
- Michael Snyder, Stanford
- Grant
- U54HG004558
- Download
- CTCF_liver_IP.png
CTCF (Homo sapiens)
neural cellsigmoid colontransverse colon
exempt from standards
- Caption
- Primary characterizations in each human tissue is not required, given the compliant characterizations in K562 and human liver as permitted by section a of page 4 of the May 2016 ENCODE antibody characterization standards document
- Submitter comment
- The tissues from GTex are limited and cannot be spared for antibody characterizations.
- Reviewer comment
- Tissues from EN-tex can be exempted from characterization with compliant characterizations in any two cell types/tissues.
- Submitted by
- Esther Chan
- Lab
- Michael Snyder, Stanford
- Grant
- U54HG006996
- Download
- ENCAB000AXU_primary.png
CTCF (Homo sapiens)
neural cell
exempt from standards
- Caption
- The ENCODE antibody standards document exempts some valuable and/or limiting samples from primary characterizations for well-characterized antibodies. They are given exemptions so that the samples can be conserved to carry out the downstream experiments.
- Submitter comment
- We do not have any of the H1-derived neurons that were distributed to the consortium for analysis in ENCODE2 left.
- Reviewer comment
- This cell type was exempted from primary characterization of antibody ENCAB000AIT by the ENCODE antibody review panel on October 17, 2016
- Submitted by
- Jessika Adrian
- Lab
- Michael Snyder, Stanford
- Grant
- U54HG006996
- Download
- H1neuron_primary.png
CTCF (Homo sapiens)
Method: immunoblot
not reviewed
- Caption
- Western blot analysis of immunoprecipitated proteins from Human Liver nuclear extracts detected using a CTCF'(07_729) antibody from Millipore. Expected protein band 120kD. Input: Input lysate, FT: Flowthrough from IP, IP: Immunoprecipitated fraction from 07_729, IgG:Control'Immunoprecipitation using IgG.
- Submitted by
- Michael Snyder
- Lab
- Michael Snyder, Stanford
- Grant
- U54HG004558
CTCF (Homo sapiens)
SK-N-SHHepG2K562
exempt from standards
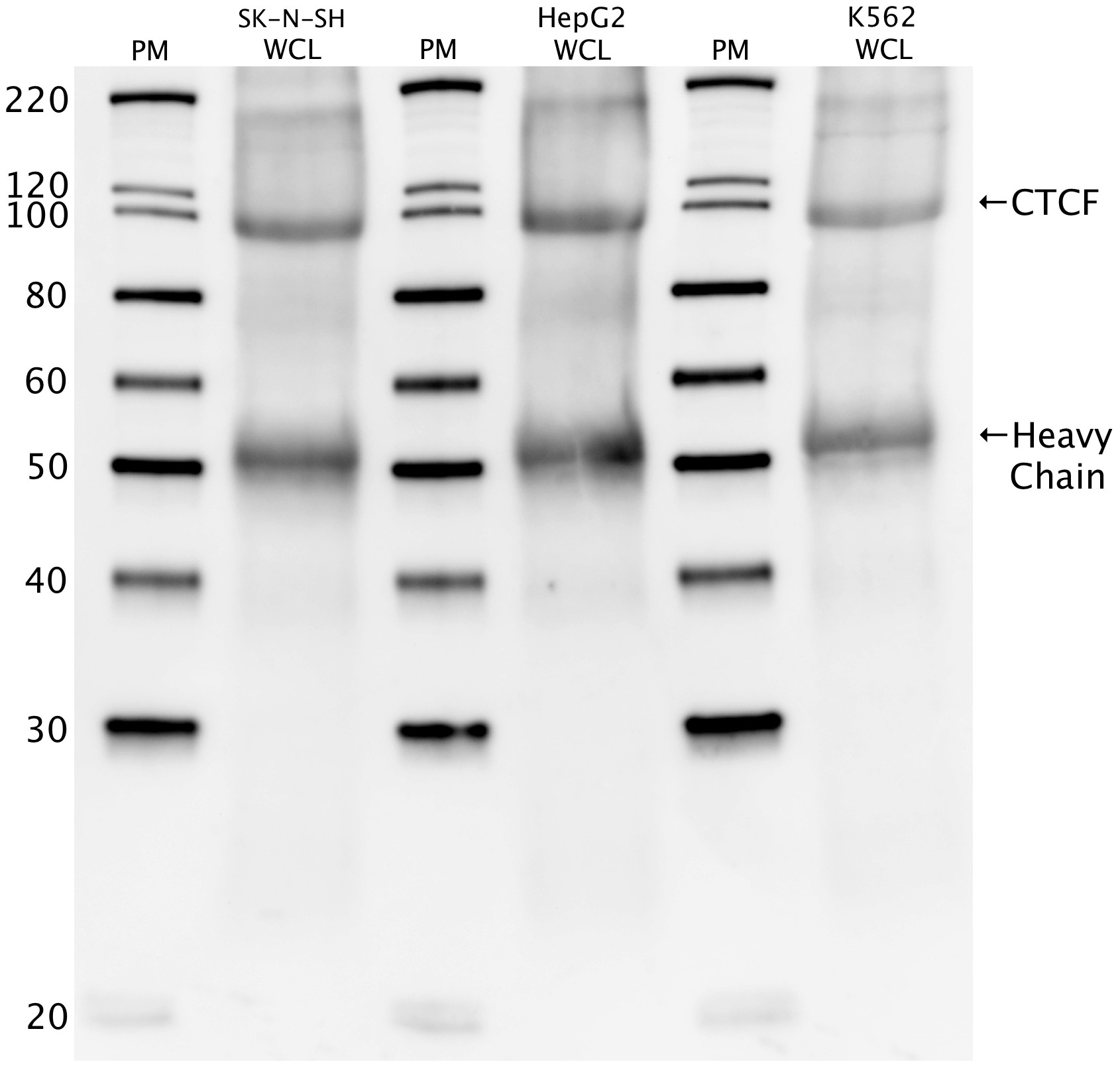
- Caption
- Western blot protocol: Whole cell lysates were immunoprecipitated using primary antibody (07-729), and the IP fraction was loaded on a 12% acrylamide gel and separated with a Bio-Rad PROTEAN II xi system. After separation, the samples were transferred to a nitrocellulose membrane with an Invitrogen iBlot system. Blotting with primary (same as that used for IP) and secondary HRP-conjugated antibodies was performed on an Invitrogen BenchPro 4100 system. Visualization was achieved using SuperSignal West Femto solution Analysis (Thermo Scientific). Results: Band of expected size visualized, representing strongest signal in the lane. Figure legend: IP-western with 07-729 in WCL (whole cell lysates) of SK-N-SH, HepG2, and K562; PM=protein marker. CTCF band is indicated, as is heavy chain of IgG.
- Submitter comment
- --
- Reviewer comment
- The antibody review panel will pass (with exemption) any immunoprecipitation characterization that meets all standards for ENCODE2. ENCODE2 did not require an IgG control
- Submitted by
- Richard Myers
- Lab
- Richard Myers, HAIB
- Grant
- U54HG004576
- Download
- human_CTCF_validation_Myers-000.png
CTCF (Homo sapiens)
not reviewed
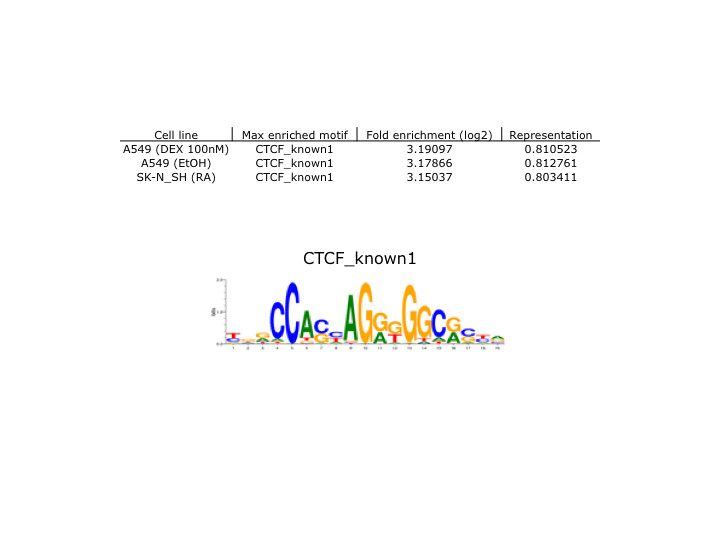
- Caption
- Motif enrichment validation protocol: Motif enrichment validation was performed by Pouya Kheradpour. Methods and results are available at http://www.broadinstitute.org/~pouyak/encode-motif-disc/. Analysis: Briefly, motif identification was performed on 100 bp peaks (50 bp on either side of peak summit), passing IDR between replicate experiments at < .01. Identified motifs were matched to known motifs for the tested TF (correlation > .75). ENCODE standards call for > 4-fold enrichment and > 10% motif representation in order to consider the identified motif validated. These standards are met and represented in the attached file, which includes cell line tested, name of maximally enriched motif, fold-enrichment value (as log2), and representation percentage. Additionally, known motif consensus sequences which were confirmed experimentally are displayed.
- Submitted by
- Richard Myers
- Lab
- Richard Myers, HAIB
- Grant
- U54HG004576
- Download
- human_CTCF_validation_Myers-001.png
CTCF (Homo sapiens)
Method: immunoblot
not reviewed
- Caption
- IP-western with 07-729 in WCL (whole cell lysates) of SK-N-SH, HepG2, and K562; PM=protein marker. CTCF band is indicated, as is heavy chain of IgG.
- Submitted by
- Richard Myers
- Lab
- Richard Myers, HAIB
- Grant
- U54HG004576
- Download
- human_CTCF_validation_Myers.pdf