Data Organization
Project overview | Using the portal | REST API | Data organization | Data submission | Encyclopedia
Metadata
The ENCODE DCC, in collaboration with the labs performing the assays and the Data Analysis Center (DAC), have defined a set of metadata used to help describe the experimental conditions that were used to generate the data, processing steps that were performed to analyze and interpret the data, and metrics to evaluate the quality and reproducibility of the data [1]. These metadata are displayed on the pages that describe the assays, biosamples, and antibodies.
Data model
Metadata for experimental assays and computational analyses are organized as JSON objects. Each object represents a specific component of the assay or analysis. A schema profile for each object type outlines the set of properties for a given object of that type. For example, an assay is represented by an Experiment object, the samples used for that assay by Biosample objects, and raw and processed data files by File objects. For a complete list of these schemas, click "Materials & Methods" -> "Schemas" at the top of any page.
A property for a given object can “link” or “point” to another object on the ENCODE Portal. For example, an Experiment object has the property replicates, a list containing the Replicate objects associated with that assay. The properties of each Replicate object are embedded directly in the experiment JSON. Each Replicate has properties of additional objects embedded within it as well, including the Library, Biosample, and Donor objects associated with that Replicate (Figure 1). These object relationships form the basis of the core data model for the ENCODE Portal (Figure 2). For details on viewing the JSON of objects on the Portal and accessing properties of objects programmatically, please see the introduction to our REST API.
{
replicates:[
{
biological_replicate_number: 1,
status: "released",
library: {
nucleic_acid_term_name: "RNA",
size_range: ">200",
biosample: {
biosample_ontology: {...},
treatments: [...],
donor: {
age: 15,
sex: "female"
}
}
}
},
{...}
]
}
Figure 1. Embedded properties of an example experiment JSON describing associated replicate, library, biosample, and donor objects.
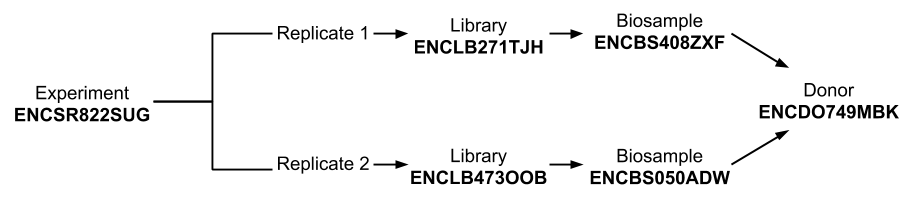
Figure 2. Representation of the core of the ENCODE DCC data model. Every experiment has at least one biological replicate.
The entire data model is available at the ENCODE DCC GitHub schema repository and visualized in svg. Replace the object name in the profile URL to view the formatted schema.
Accessions
The DCC creates accessions for metadata that can be reused in experimental protocols and computational analyses. This practice ensures that the exact assay or reagent is being referred to when assays are being discussed or files are being analyzed. The accessions follow the format ENC[SR|BS|DO|GM|AB|LB|FF|PL][0-9]{3}[A-Z]{3} where [SR|BS|DO|GM|AB|LB|FF|PL] refer to the metadata type given the accession. This format is ideal for referring to ENCODE data in publications. Accessions are given to the following types of metadata:
- An assay (ENCSR): Each assay is given an accession. Assays may contain one or more biological replicates. Typically, replicates of a given assay will be performed using the same method and biosample type, and if applicable will investigate the same target. Each collection of assays, such as a time series or reference epigenome, is also given an ENCSR accession. A sample accession for an assay is ENCSR535GFO.
- A biosample (ENCBS): An biosample accession refers to a tube or sample of that biological material used in an assay. For example, the following would each be given a biosample accession: (1) a batch of a cell line grown on a specific day, (2) the isolation of a primary cell culture on a specific day, or (3) the dissection of a tissue sample on a specific day. A sample accession for a biosample is ENCBS646JZQ.
- A strain or donor (ENCDO): Every strain (for model organisms) and donor (for humans) is given a donor accession. This accession allows multiple biosamples obtained from a single donor to be grouped together. A sample accession for a donor is ENCDO451RUA.
- A genetic modification (ENCGM): If a biosample or donor is genetically altered in some way, the applied modification is given an accession. This accession allows multiple biosamples or donors with the same genetic modification to be grouped together. A sample accession for a genetic modification is ENCGM815ANE.
- An antibody lot (ENCAB): Each unique antibody lot is given an accession so that assays can refer specifically to that antibody. Each antibody lot is also associated with characterizations for its target in a species. A sample accession for an antibody lot is ENCAB615WUN.
- A library (ENCLB): Each unique library that can be resequenced is given an accession. This accession ensures the correct files are associated with the nucleic acid material created from the biosample. A sample accession for a library is ENCLB829EXR.
- A file (ENCFF): Each data file available for download is given an accession. Each assay contains a list of file accessions belonging to that assay. The file accession and file format are included in the name of the file itself. A sample accession for a file is ENCFF437QEP.
- A pipeline (ENCPL): Each data processing pipeline is given an accession. This accession groups together all information regarding file input, file output, and software used at each step of the pipeline. A sample accession for a pipeline is ENCPL272XAE.
Ontologies
The DCC uses ontologies to annotate experimental metadata in order to provide improved searching capabilities across the data, to build a technical framework for ensuring the accuracy of the metadata, and to facilitate the interoperability with other genomic databases. Ontologies provide consistent language, which facilitates the integration of datasets from diverse projects and is essential to ensure all the correct results are returned when browsing or searching the metadata. The relations between ontology terms are used by the DCC in order to group samples and experiments by higher-level terms. For example, selecting brain on the Organ facet will return Experiments conducted on tissue from any region of the brain, all primary brain cells, and all cell lines derived from brain cells. The Cell facet also utilizes the ontological relations between terms in a similar manner.
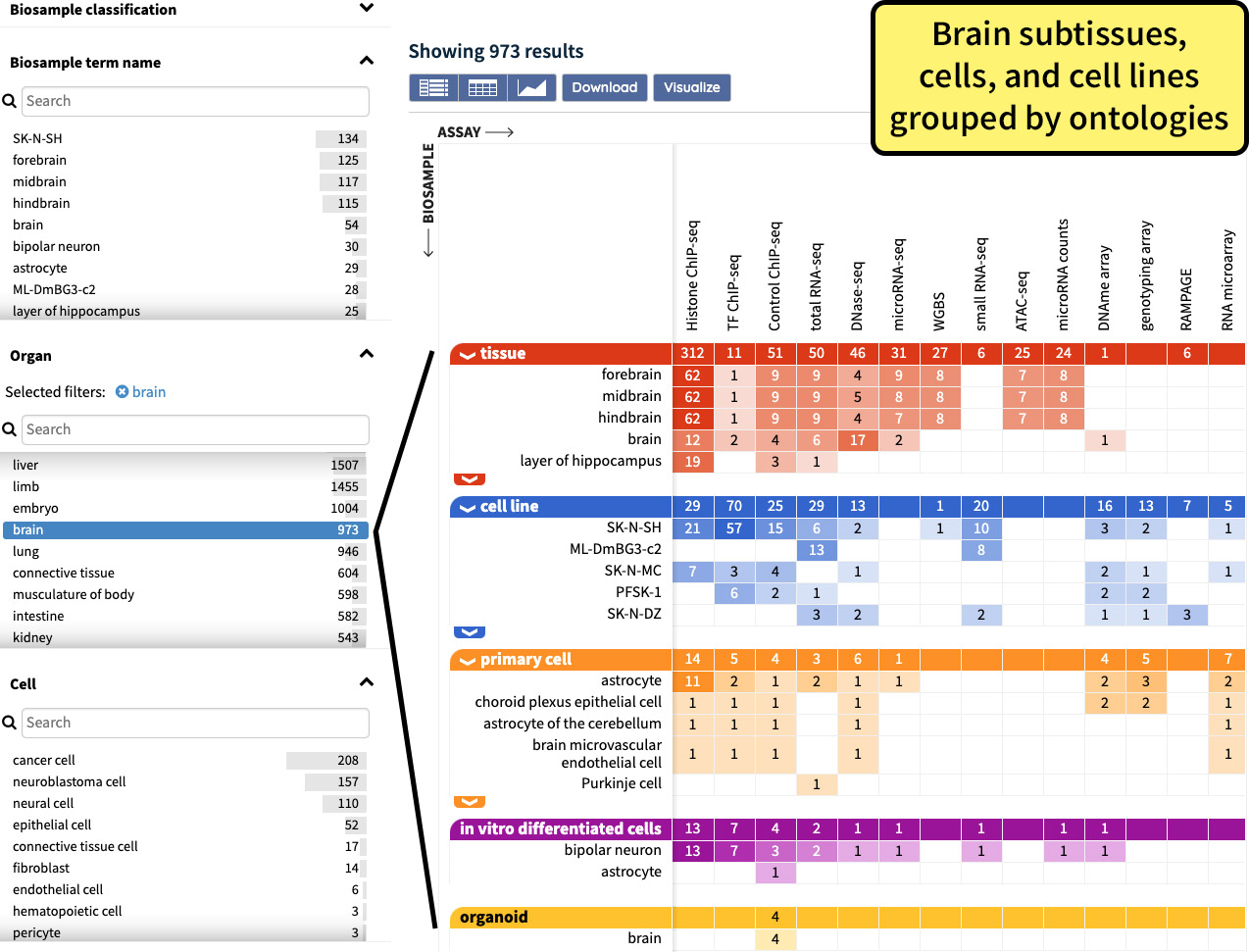
Figure 3. The selection of brain on the Organ facet finds all assays performed on multiple biosample types and regions of the brain.
Additionally, the DCC imports synonyms from each ontology term allowing for flexibility in free text search terms, while maintaining the controlled description of the metadata via the official term name. In the process, we are contributing to community efforts to improve these ontologies by working with ontology databases for New Term Requests and improving annotations of existing terms. To this end, the ENCODE DCC is using the following ontologies to capture specific metadata categories:
- Tissues: Uber Anatomy Ontology (UBERON)
- Primary cells: Cell Ontology (CL)
- Cell lines: Experimental Factor Ontology (EFO) and Cell Line Ontology (CLO)
- Experimental assays (such as RNA-seq, ChIP-seq): Ontology for Biomedical Investigations (OBI)
- Chemical treatments (such as estradiol, ethanol): Chemical Entities of Biological Interest (ChEBI)
- Nucleic acid being sequenced (such as microRNA, mRNA): Sequence Ontology (SO)
- Target categories: Gene Ontology (GO)
- Biosample disease status: Disease Ontology (DO)
The ENCODE DCC GitHub repository has details regarding the generation and updating of ontologies on the ENCODE Portal.
Dependencies and Audits
In order to ensure metadata accuracy, each schema has a set of dependencies to enforce proper modeling when related metadata are submitted. For example, if a FASTQ file is posted, a dependency requires an additional property indicating whether that file is single-ended or paired-ended. If paired-ended is selected, an additional dependency requires the ID of the paired file is included as well. These safeguards ensure that the appropriate metadata are in place prior to the submission of a new object.
After submission, a system of audits are used to identify inconsistencies in the data. While dependencies enforce metadata within an object, audits typically check metadata between linked objects. For example, these data audits include checks that ensure that a ChIP-seq Experiment lists control Experiments with matching biosample metadata and that spike-ins are used for RNA-seq assays. These audits are also used to communicate details of ENCODE data, such as data quality relative to standards, to the public. Each audit is designated a color depending on its severity, and is displayed on the search page and individual object pages. For a comprehensive list and description of the various audits implemented on the Portal, see the Audits page.
References
[1] Hong EL, Sloan CA, Chan ET, Davidson JM, Malladi VS, Strattan JS, Hitz BC, Gabdank I, Narayanan AK, Ho M, Lee BT, Rowe LD, Dreszer TR, Roe GR, Podduturi NR, Tanaka F, Hilton JA, Cherry JM. Principles of metadata organization at the ENCODE coordination center. Database. 2016 Mar;2016:1-10. PMID: 26980513