ENCAB000AJM
Antibody against Homo sapiens EP300, Mus musculus EP300
Homo sapiens
HeLa-S3, K562, GM12878, HepG2, neural cell
characterized to standards with exemption
Mus musculus
liver, lung, midbrain, hindbrain, heart, kidney, stomach, forebrain, intestine, limb
characterized to standards with exemption
Homo sapiens
any cell type or tissue
partially characterized
Mus musculus
any cell type or tissue
partially characterized
- Status
- released
- Source (vendor)
- Santa Cruz Biotech
- Product ID
- sc-585
- Lot ID
- D0411
- Characterized targets
- EP300 (Homo sapiens), EP300 (Mus musculus)
- Host
- rabbit
- Clonality
- polyclonal
- External resources
Characterizations
EP300 (Homo sapiens)
not reviewed
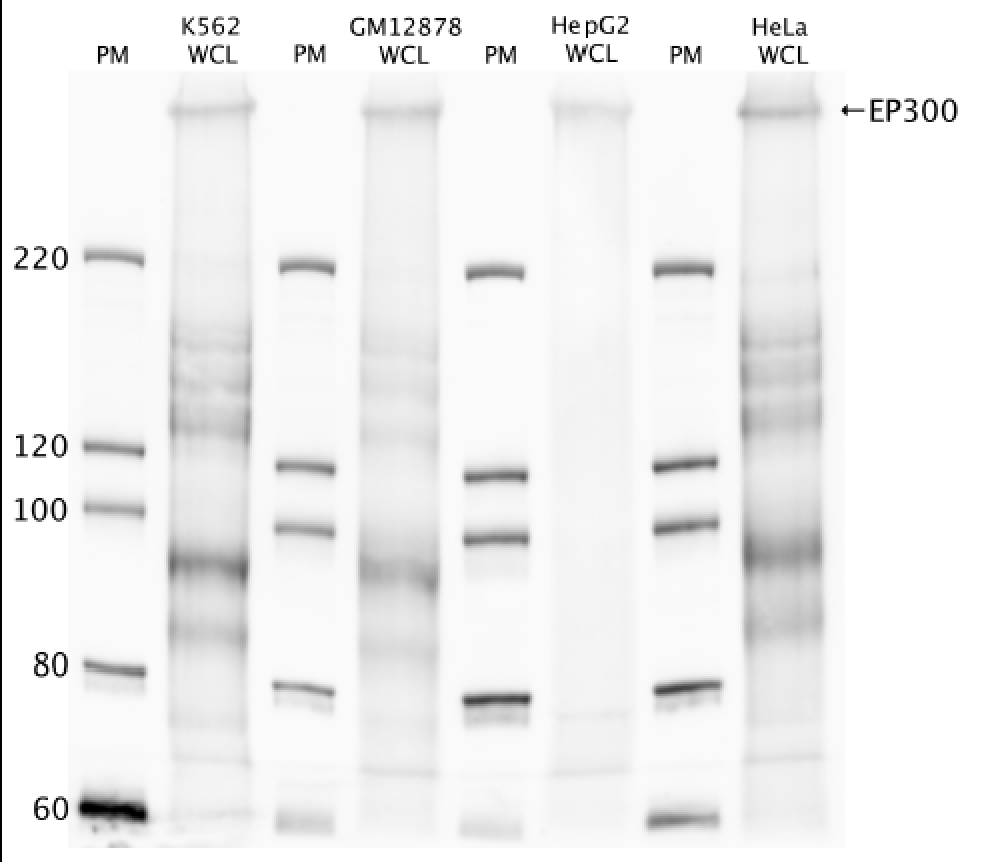
- Caption
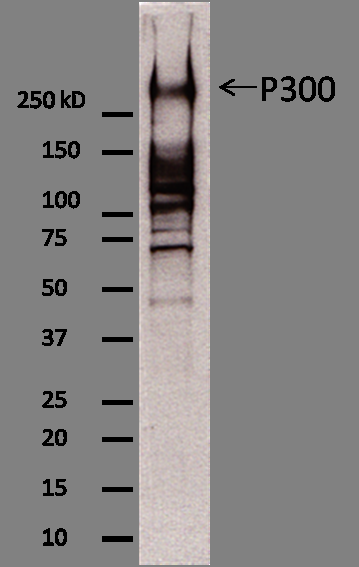
- Western blot protocol: Whole cell lysates were immunoprecipitated using primary antibody (sc-585), and the IP fraction was loaded on a 12% acrylamide gel and separated with a Bio-Rad PROTEAN II xi system. After separation, the samples were transferred to a nitrocellulose membrane with an Invitrogen iBlot system. Blotting with primary (same as that used for IP) and secondary HRP-conjugated antibodies was performed on an Invitrogen BenchPro 4100 system. Visualization was achieved using SuperSignal West Femto solution (Thermo Scientific). Results: Band of expected size visualized, representing strongest signal in the lane. Figure legend: IP-western with sc-585 in WCL (whole cell lysates) of K562, GM12878, HepG2, and HeLa; PM=protein marker. p300 band is indicated.
- Submitted by
- Richard Myers
- Lab
- Richard Myers, HAIB
- Grant
- U54HG004576
- Documents
EP300 (Homo sapiens)
compliant
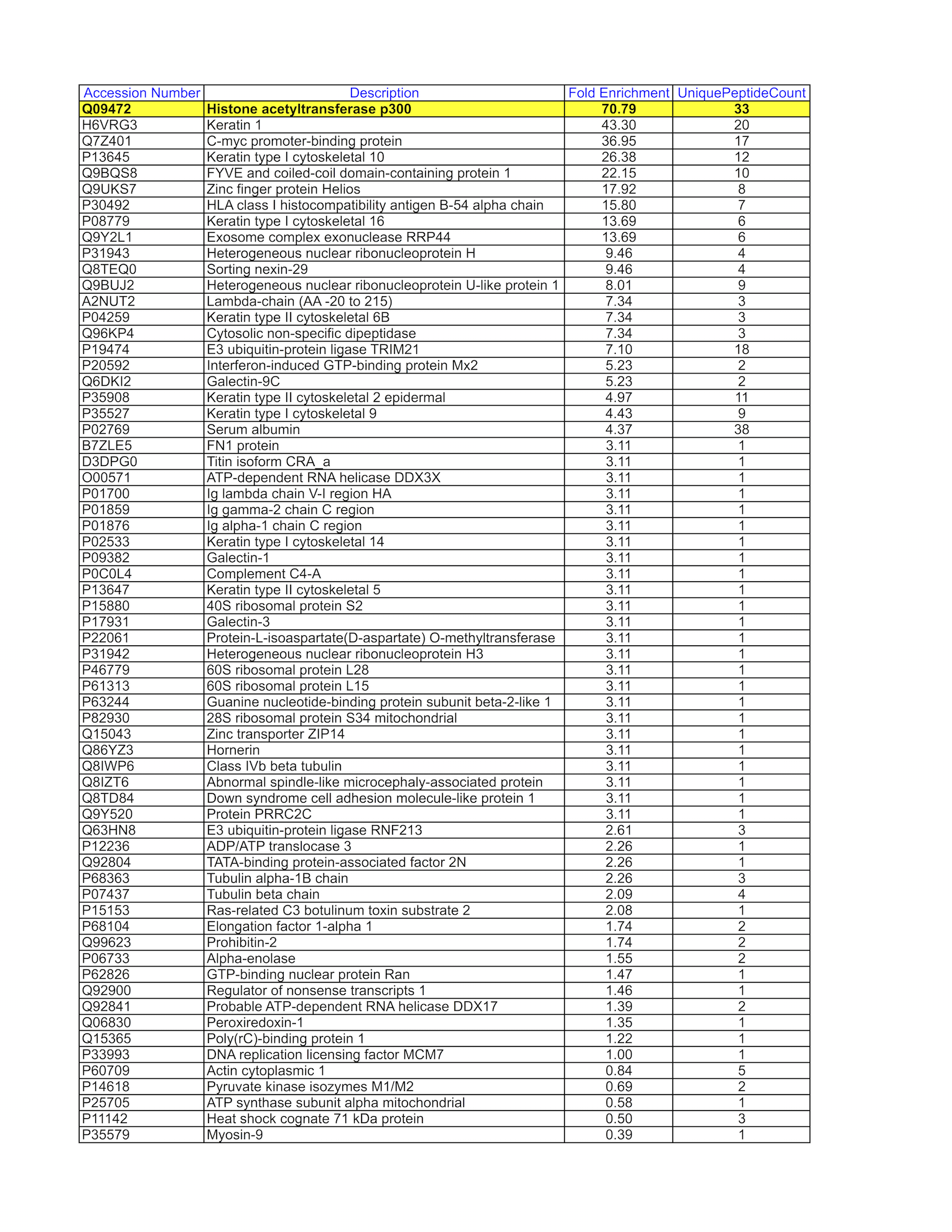
- Caption
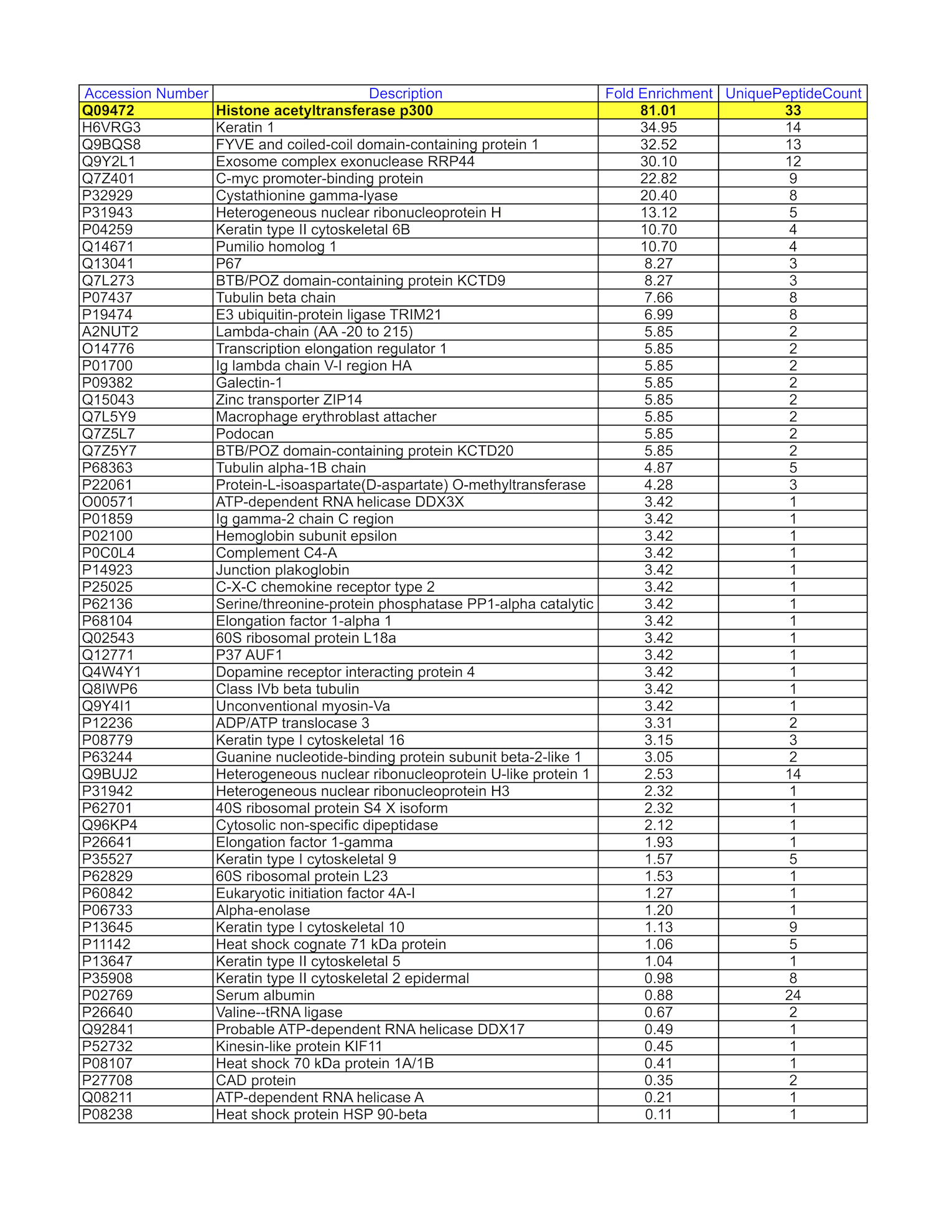
- GM12878 whole cell lysate was immunoprecipitated using the primary antibody (Santa Cruz Biotechnology; sc-585). The IP fraction was loaded on a 12% Bio-Rad TGX gel and separated with the Bio-Rad Tetra Cell system. The whole lane was excised and sent to the University of Alabama at Birmingham Cancer Center Mass Spectrometry/Proteomics Shared Facility. Analysis of whole lane from GM12878: The sample was analyzed on a LTQ XL Linear Ion Trap Mass Spectrometer by LC-ESI-MS/MS. Peptides were identified using SEQUEST tandem mass spectral analysis with probability based matching at p < 0.05. SEQUEST results were reported with ProteinProphet protXML Viewer (TPP v4.4 JETSTREAM) and filtered for a minimum probability of 0.9. All protein hits that met these criteria were reported, including common contaminants. Fold enrichment for each protein reported was determined using a custom script based on the FC-B score calculation from the reference Mellacheruvu et al., 2013. The CRAPome: a contaminant repository for affinity purification mass spectrometry data. Nat. Methods. 10(8):730-736. Doi:10.1038/nmeth.2557. The target protein, EP300, was identified as the 1st enriched protein and the 1st transcription factor based on IP-Mass Spectrometry.
- Submitter comment
- In some cases, we exercised the option of extracting the whole lane to send for IP-mass spectrometry as opposed to cutting and sending discrete bands from a gel. This is especially the case when the antibody results in multiple and non-discrete bands or no visible bands in the predicted size range of the protein when observed by IP-Western. To ensure that the targeted protein can be identified in the IP, whole cell lysates are loaded in a 12% Bio-Rad TGX gel and run for 5-10 minutes using the Bio-Rad Tetra Cell system. The entire gel lane or just the single unseparated band is excised from the gel and sent for analysis. We do not provide coomassie blue-stained gel images in these instances as all one observes is a thick, unresolved band near the loading well.
- Submitted by
- Mark Mackiewicz
- Lab
- Richard Myers, HAIB
- Grant
- U54HG006998
- Download
- EP300_GM12878-WL.png
EP300 (Homo sapiens)
compliant
- Caption
- HepG2 whole cell lysate was immunoprecipitated using the primary antibody (Santa Cruz Biotechnology; sc-585). The IP fraction was loaded on a 12% Bio-Rad TGX gel and separated with the Bio-Rad Tetra Cell system. The whole lane was excised and sent to the University of Alabama at Birmingham Cancer Center Mass Spectrometry/Proteomics Shared Facility. Analysis of whole lane from HepG2: The sample was analyzed on a LTQ XL Linear Ion Trap Mass Spectrometer by LC-ESI-MS/MS. Peptides were identified using SEQUEST tandem mass spectral analysis with probability based matching at p < 0.05. SEQUEST results were reported with ProteinProphet protXML Viewer (TPP v4.4 JETSTREAM) and filtered for a minimum probability of 0.9. All protein hits that met these criteria were reported, including common contaminants. Fold enrichment for each protein reported was determined using a custom script based on the FC-B score calculation from the reference Mellacheruvu et al., 2013. The CRAPome: a contaminant repository for affinity purification mass spectrometry data. Nat. Methods. 10(8):730-736. Doi:10.1038/nmeth.2557. The target protein, EP300, was identified as the 2nd enriched protein and the 1st transcription factor based on IP-Mass Spectrometry.
- Submitter comment
- In some cases, we exercised the option of extracting the whole lane to send for IP-mass spectrometry as opposed to cutting and sending discrete bands from a gel. This is especially the case when the antibody results in multiple and non-discrete bands or no visible bands in the predicted size range of the protein when observed by IP-Western. To ensure that the targeted protein can be identified in the IP, whole cell lysates are loaded in a 12% Bio-Rad TGX gel and run for 5-10 minutes using the Bio-Rad Tetra Cell system. The entire gel lane or just the single unseparated band is excised from the gel and sent for analysis. We do not provide coomassie blue-stained gel images in these instances as all one observes is a thick, unresolved band near the loading well.
- Submitted by
- Mark Mackiewicz
- Lab
- Richard Myers, HAIB
- Grant
- U54HG006998
- Download
- EP300_HepG2-WL.png
EP300 (Homo sapiens)
K562GM12878HepG2
exempt from standards
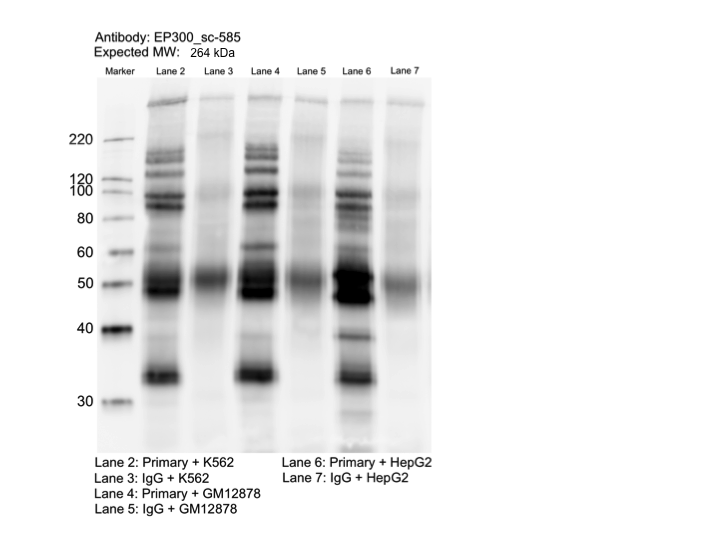
- Caption
- Whole cell lysates of K562, GM12878, and HepG2 were immunoprecipitated using the primary antibody (Santa Cruz Biotechnology; sc-585). The IP fraction was separated on a 12% acrylamide gel with the Bio-Rad PROTEAN II xi system. After separation, the samples were transferred to a nitrocellulose membrane with an Invitrogen iBlot system. The membrane was probed with the primary antibody (same as that used for IP) and a secondary HRP-conjugated antibody. The resulting bands were visualized with SuperSignal West Femto Solution (Thermo Scientific). Protein Marker (PM) is labeled in kDa. The approximate size of EP300 is ~264 kDa, although no distinguishable band was observed at this size range.
- Submitter comment
- --
- Reviewer comment
- Band of interest is not 50% of overall signal in lane. Rescued by mass spectrometry analysis
- Submitted by
- Mark Mackiewicz
- Lab
- Richard Myers, HAIB
- Grant
- U54HG006998
- Download
- EP300_IP-WB.png
EP300 (Homo sapiens)
compliant
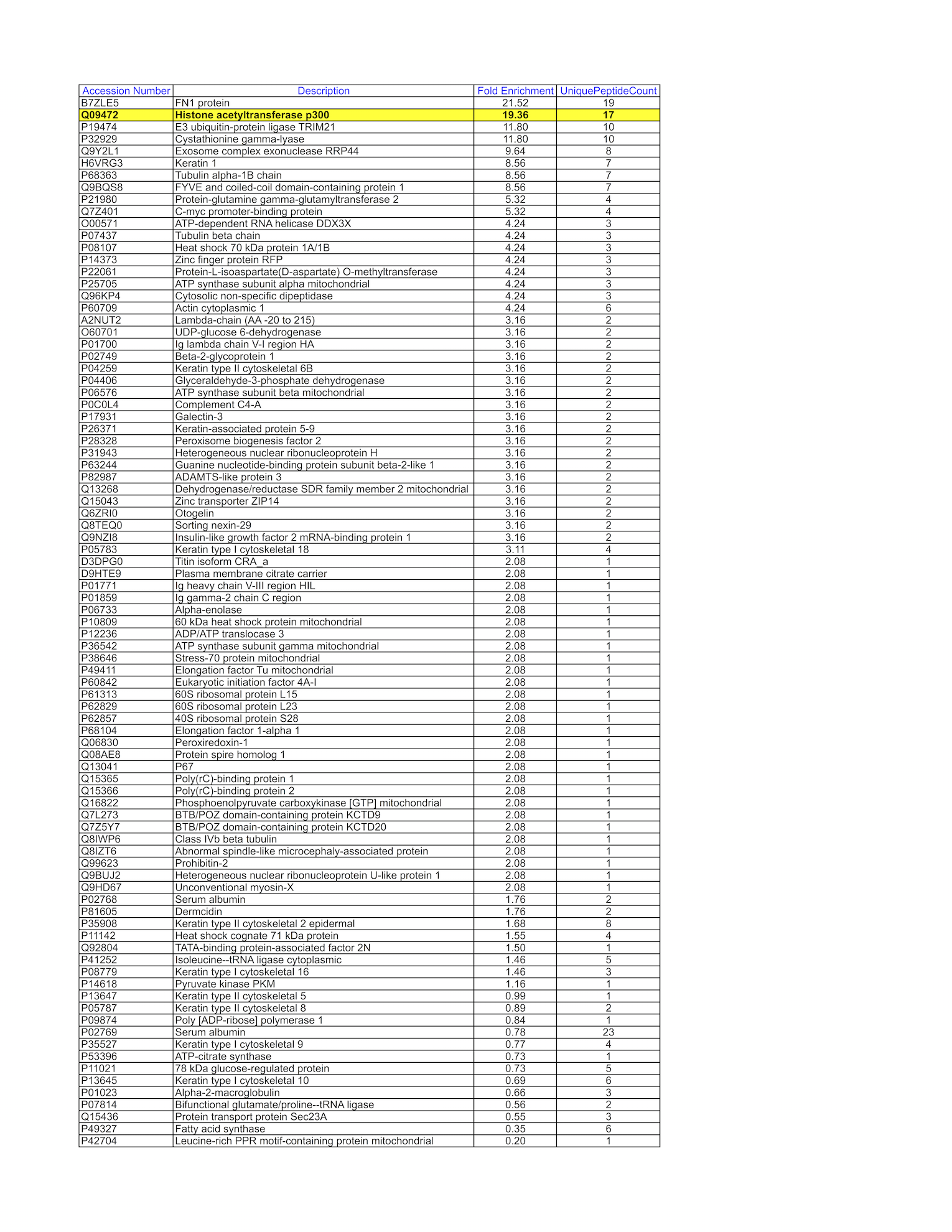
- Caption
- K562 whole cell lysate was immunoprecipitated using the primary antibody (Santa Cruz Biotechnology; sc-585). The IP fraction was loaded on a 12% Bio-Rad TGX gel and separated with the Bio-Rad Tetra Cell system. The whole lane was excised and sent to the University of Alabama at Birmingham Cancer Center Mass Spectrometry/Proteomics Shared Facility. Analysis of whole lane from K562: The sample was analyzed on a LTQ XL Linear Ion Trap Mass Spectrometer by LC-ESI-MS/MS. Peptides were identified using SEQUEST tandem mass spectral analysis with probability based matching at p < 0.05. SEQUEST results were reported with ProteinProphet protXML Viewer (TPP v4.4 JETSTREAM) and filtered for a minimum probability of 0.9. All protein hits that met these criteria were reported, including common contaminants. Fold enrichment for each protein reported was determined using a custom script based on the FC-B score calculation from the reference Mellacheruvu et al., 2013. The CRAPome: a contaminant repository for affinity purification mass spectrometry data. Nat. Methods. 10(8):730-736. Doi:10.1038/nmeth.2557. The target protein, EP300, was identified as the 1st enriched protein and the 1st transcription factor based on IP-Mass Spectrometry.
- Submitter comment
- In some cases, we exercised the option of extracting the whole lane to send for IP-mass spectrometry as opposed to cutting and sending discrete bands from a gel. This is especially the case when the antibody results in multiple and non-discrete bands or no visible bands in the predicted size range of the protein when observed by IP-Western. To ensure that the targeted protein can be identified in the IP, whole cell lysates are loaded in a 12% Bio-Rad TGX gel and run for 5-10 minutes using the Bio-Rad Tetra Cell system. The entire gel lane or just the single unseparated band is excised from the gel and sent for analysis. We do not provide coomassie blue-stained gel images in these instances as all one observes is a thick, unresolved band near the loading well.
- Submitted by
- Mark Mackiewicz
- Lab
- Richard Myers, HAIB
- Grant
- U54HG006998
- Download
- EP300_K562-WL.png
EP300 (Mus musculus)
HeLa-S3
exempt from standards
- Caption
- The ENCODE antibody standards document exempts some valuable and/or limiting samples from primary characterizations for well-characterized antibodies. They are given exemptions so that the samples can be conserved to carry out the downstream experiments.
- Submitter comment
- Submitting for Peggy Farnham to affect Snyder experiments.
- Reviewer comment
- Submitting for Peggy Farnham to affect Snyder experiments.
- Submitted by
- Kathrina Onate
- Lab
- Michael Snyder, Stanford
- Grant
- UM1HG009442
- Download
- exempted.png
EP300 (Homo sapiens)
neural cell
exempt from standards
- Caption
- The ENCODE antibody standards document exempts some valuable and/or limiting samples from primary characterizations for well-characterized antibodies. They are given exemptions so that the samples can be conserved to carry out the downstream experiments.
- Submitter comment
- We do not have any of the H1-derived neurons that were distributed to the consortium for analysis in ENCODE2 left.
- Reviewer comment
- This cell type was exempted from primary characterization of antibody ENCAB000AIT by the ENCODE antibody review panel on October 17, 2016
- Submitted by
- Jessika Adrian
- Lab
- Michael Snyder, Stanford
- Grant
- U54HG006996
- Download
- H1neuron_primary.png
EP300 (Mus musculus)
not reviewed
- Caption
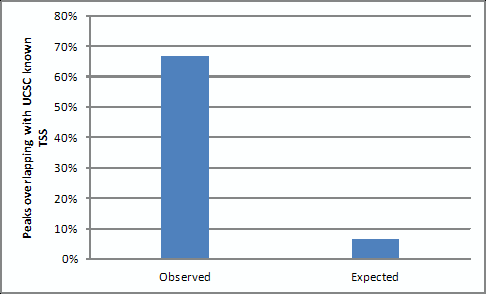
- We performed ChIP-seq of P300 in mESCs with two biological replicates, and called peaks by MACS with the default parameter (P value < 1e-5). 67% out of the 16,054 distal P300 binding sites overlap with predicted enhancers by H3K4me1, compared to random expected 6.5% (P < 2.2 x e -16 (chi-square test). This result shows that the P300 antibody we use is specific.
- Submitted by
- Yin Shen
- Lab
- Bing Ren, UCSD
- Grant
- R01HG003991
EP300 (Mus musculus)
liverlungmidbrainhindbrainheartkidneystomachforebrainintestinelimb
exempt from standards
- Caption
- The ENCODE Binding Working Group finds for some valuable tissues that recreating a primary on well characterized antibodies is not cost effective. Therefore, they allow exemption from standards for these tissues.
- Submitter comment
- The lab is asking for an exemption for several tissue cells due to the lack of resource to make a primary characterization for them
- Reviewer comment
- Exempted by the Feb 29, 2016 antibody review panel
- Submitted by
- Mark Mackiewicz
- Lab
- Richard Myers, HAIB
- Grant
- U54HG006998
- Download
- No_tissue.png