ENCAB725LFJ
Antibody against Homo sapiens CBFB
Homo sapiens
K562, GM12878, HepG2, MCF-7
characterized to standards with exemption
- Status
- released
- Source (vendor)
- GeneTex
- Product ID
- GTX106820
- Lot ID
- 39981
- Characterized targets
- CBFB (Homo sapiens)
- Host
- rabbit
- Clonality
- polyclonal
- Purification
- affinity
- Isotype
- IgG
- Antigen description
- Recomb fragment corresp to a region within amino acids 1 and 182 of PEBP2beta (Uniprot ID#Q13951)
- External resources
Characterizations
CBFB (Homo sapiens)
compliant
- Caption
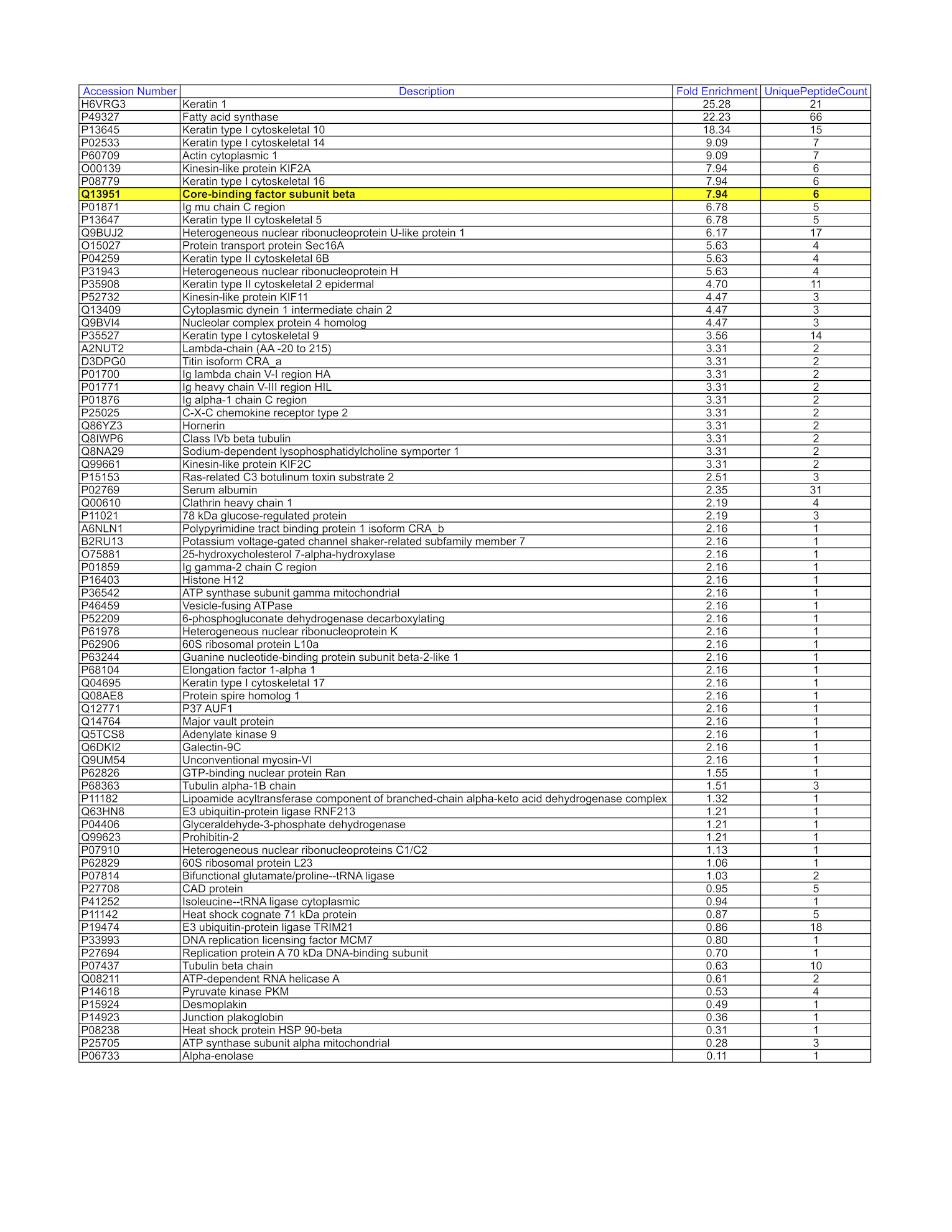
- GM12878 whole cell lysate was immunoprecipitated using the primary antibody (GeneTex; GTX106820). The IP fraction was loaded on a 12% Bio-Rad TGX gel and separated with the Bio-Rad Tetra Cell system. The whole lane was excised and sent to the University of Alabama at Birmingham Cancer Center Mass Spectrometry/Proteomics Shared Facility. Analysis of whole lane from GM12878: The sample was analyzed on a LTQ XL Linear Ion Trap Mass Spectrometer by LC-ESI-MS/MS. Peptides were identified using SEQUEST tandem mass spectral analysis with probability based matching at p < 0.05. SEQUEST results were reported with ProteinProphet protXML Viewer (TPP v4.4 JETSTREAM) and filtered for a minimum probability of 0.9. All protein hits that met these criteria were reported, including common contaminants. Fold enrichment for each protein reported was determined using a custom script based on the FC-B score calculation from the reference Mellacheruvu et al., 2013. The CRAPome: a contaminant repository for affinity purification mass spectrometry data. Nat. Methods. 10(8):730-736. Doi:10.1038/nmeth.2557. The target protein, CBFB, was identified as the 8th enriched protein and the 1st transcription factor based on IP-Mass Spectrometry.
- Submitter comment
- In some cases, we exercised the option of extracting the whole lane to send for IP-mass spectrometry as opposed to cutting and sending discrete bands from a gel. This is especially the case when the antibody results in multiple and non-discrete bands or no visible bands in the predicted size range of the protein when observed by IP-Western. To ensure that the targeted protein can be identified in the IP, whole cell lysates are loaded in a 12% Bio-Rad TGX gel and run for 5-10 minutes using the Bio-Rad Tetra Cell system. The entire gel lane or just the single unseparated band is excised from the gel and sent for analysis. We do not provide coomassie blue-stained gel images in these instances as all one observes is a thick, unresolved band near the loading well.
- Submitted by
- Mark Mackiewicz
- Lab
- Richard Myers, HAIB
- Grant
- U54HG006998
- Download
- CBFB-WL.png
CBFB (Homo sapiens)
K562GM12878HepG2MCF-7
exempt from standards
- Caption
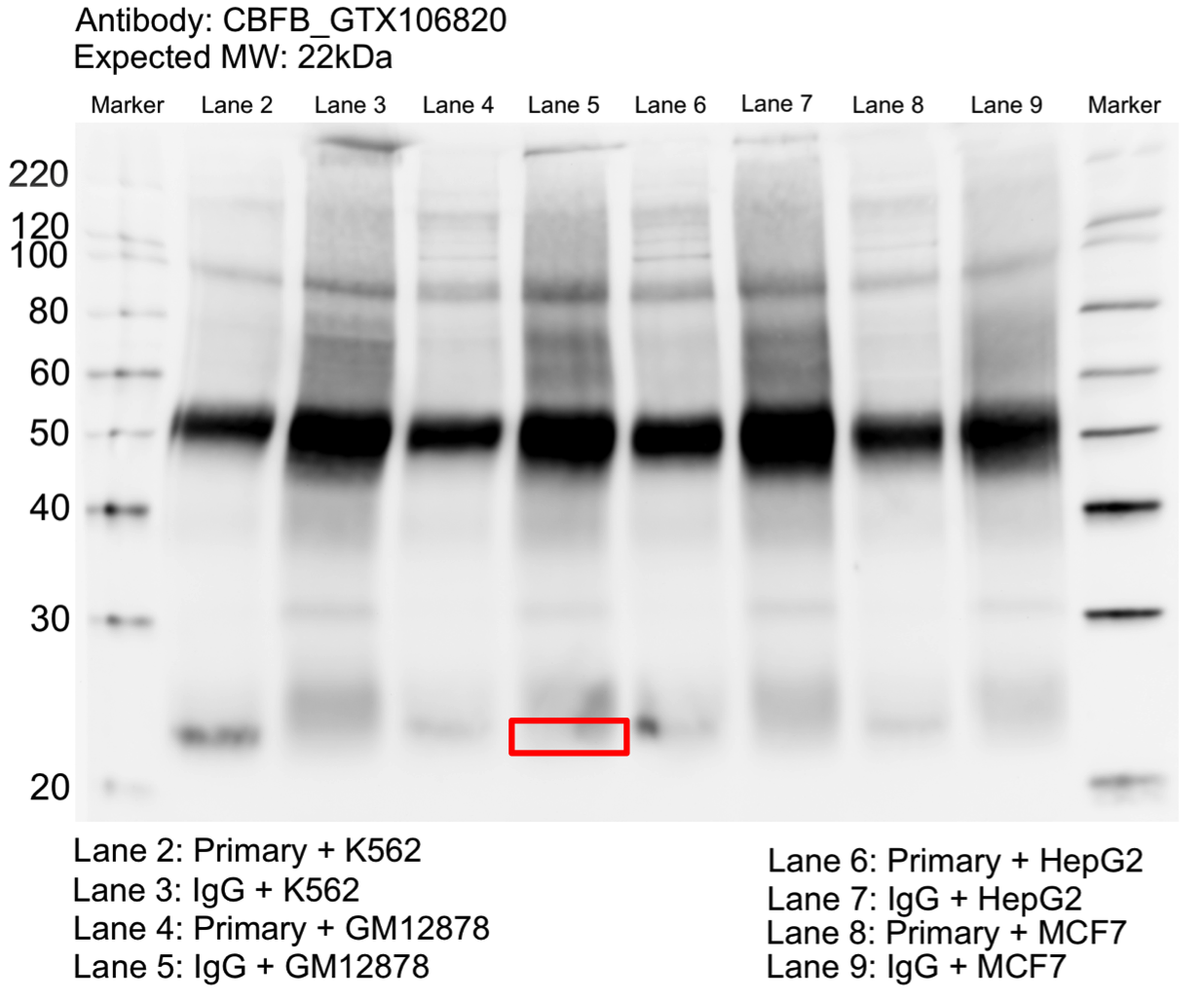
- Whole cell lysates of K562, GM12878, HepG2, and MCF7 were immunoprecipitated using the primary antibody (GeneTex; GTX106820). The IP fraction was separated on a 12% acrylamide gel with the Bio-Rad PROTEAN II xi system. After separation, the samples were transferred to a nitrocellulose membrane with an Invitrogen iBlot system. The membrane was probed with the primary antibody (same as that used for IP) and a secondary HRP-conjugated antibody. The resulting bands were visualized with SuperSignal West Femto Solution (Thermo Scientific). Protein Marker (PM) is labeled in kDa. A single prominent band was detected at ~22 kDa.
- Submitter comment
- In some cases, we exercised the option of extracting the whole lane to send for IP-mass spectrometry as opposed to cutting and sending discrete bands from a gel. This is especially the case when the antibody results in multiple and non-discrete bands or no visible bands in the predicted size range of the protein when observed by IP-Western. To ensure that the targeted protein can be identified in the IP, whole cell lysates are loaded in a 12% Bio-Rad TGX gel and run for 5-10 minutes using the Bio-Rad Tetra Cell system. The entire gel lane or just the single unseparated band is excised from the gel and sent for analysis. We do not provide coomassie blue-stained gel images in these instances as all one observes is a thick, unresolved band near the loading well.
- Reviewer comment
- Not within 20% of expected size. Rescued by mass spectrometry analysis
- Submitted by
- Mark Mackiewicz
- Lab
- Richard Myers, HAIB
- Grant
- U54HG006998
- Download
- CBFB_IP-WB.png