ENCAB661MMQ
Antibody against Homo sapiens HNF1A
Homo sapiens
HepG2
characterized to standards
- Status
- released
- Source (vendor)
- GeneTex
- Product ID
- GTX113850
- Lot ID
- 40135
- Characterized targets
- HNF1A (Homo sapiens)
- Host
- rabbit
- Clonality
- polyclonal
- Purification
- affinity
- Isotype
- IgG
- Antigen description
- Recomb fragment corresp to region within amino acids 35 and 268 of HNF1 alpha (Uniprot ID#P20823)
- External resources
Characterizations
HNF1A (Homo sapiens)
compliant
- Caption
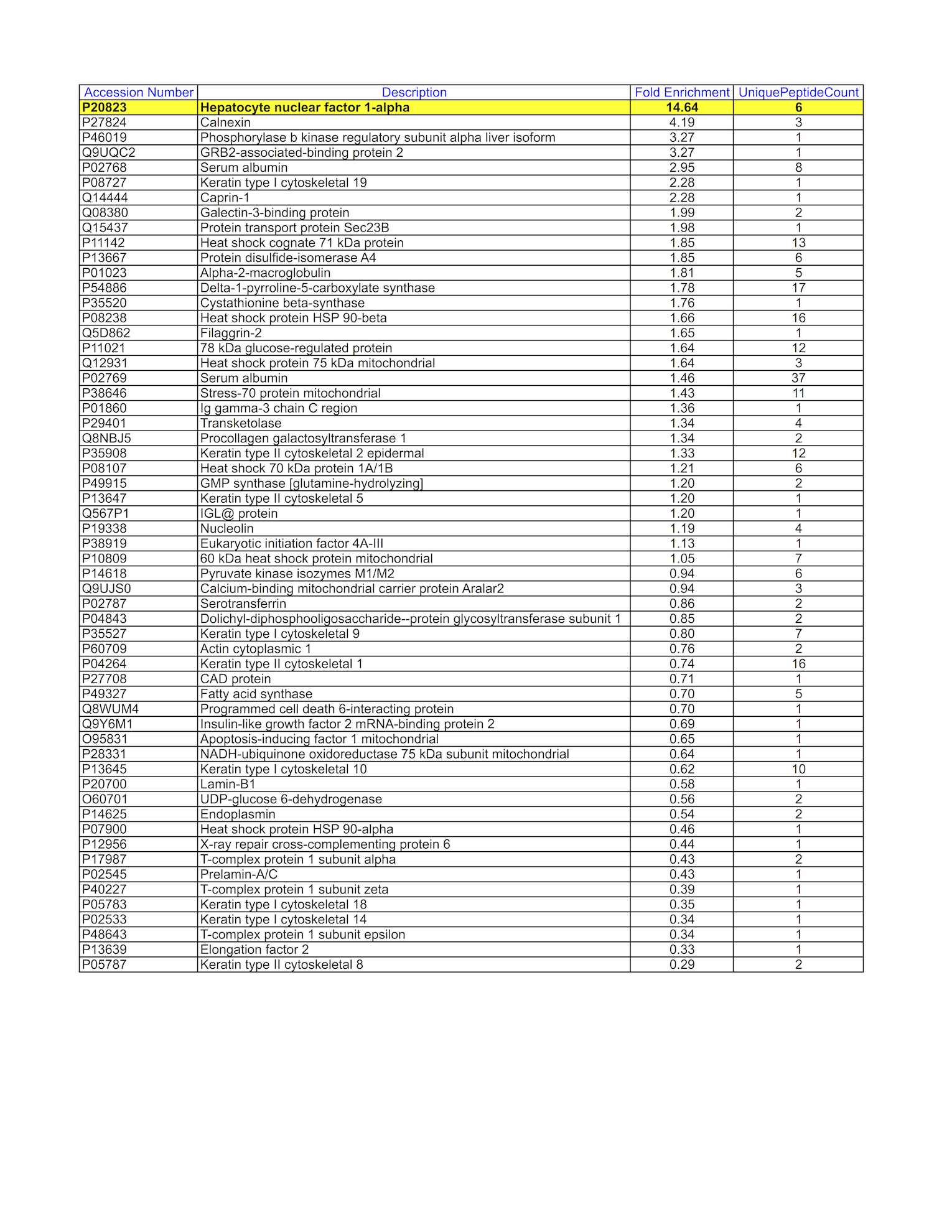
- HepG2 whole cell lysate was immunoprecipitated using the primary antibody (GeneTex; GTX113850). The IP fraction was loaded on a 12% Bio-Rad TGX gel and separated with the Bio-Rad Tetra Cell system. A gel fragment (rectangle outline) corresponding to the bands indicated on the Coomassie Blue stained gel image were excised and sent to the University of Alabama at Birmingham Cancer Center Mass Spectrometry/Proteomics Shared Facility. Analysis of gel fragment 1 from HepG2: The sample was analyzed on a LTQ XL Linear Ion Trap Mass Spectrometer by LC-ESI-MS/MS. Peptides were identified using SEQUEST tandem mass spectral analysis with probability based matching at p < 0.05. SEQUEST results were reported with ProteinProphet protXML Viewer (TPP v4.4 JETSTREAM) and filtered for a minimum probability of 0.9. All protein hits that met these criteria were reported, including common contaminants. Fold enrichment for each protein reported was determined using a custom script based on the FC-B score calculation from the reference Mellacheruvu et al., 2013. The CRAPome: a contaminant repository for affinity purification mass spectrometry data. Nat. Methods. 10(8):730-736. Doi:10.1038/nmeth.2557. The target protein, HNF1A, was identified as the 1st ranked enriched protein and the 1st ranked transcription factor based on IP-Mass Spectrometry.
- Submitted by
- Mark Mackiewicz
- Lab
- Richard Myers, HAIB
- Grant
- U54HG006998
- Download
- HNF1A_1_IP-MS.png
HNF1A (Homo sapiens)
compliant
- Caption
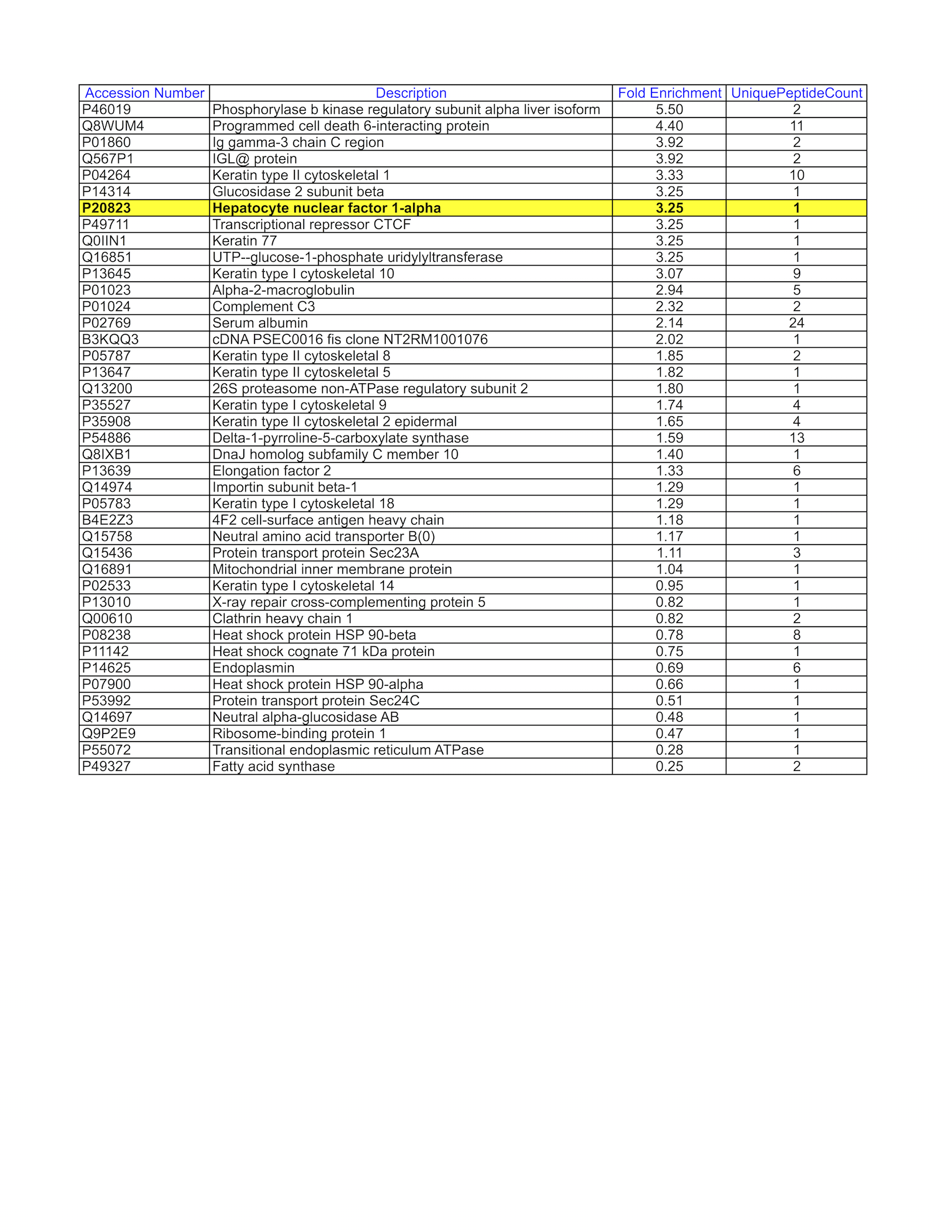
- HepG2 whole cell lysate was immunoprecipitated using the primary antibody (GeneTex; GTX113850). The IP fraction was loaded on a 12% Bio-Rad TGX gel and separated with the Bio-Rad Tetra Cell system. A gel fragment (rectangle outline) corresponding to the bands indicated on the Coomassie Blue stained gel image were excised and sent to the University of Alabama at Birmingham Cancer Center Mass Spectrometry/Proteomics Shared Facility. Analysis of gel fragment 2 from HepG2: The sample was analyzed on a LTQ XL Linear Ion Trap Mass Spectrometer by LC-ESI-MS/MS. Peptides were identified using SEQUEST tandem mass spectral analysis with probability based matching at p < 0.05. SEQUEST results were reported with ProteinProphet protXML Viewer (TPP v4.4 JETSTREAM) and filtered for a minimum probability of 0.9. All protein hits that met these criteria were reported, including common contaminants. Fold enrichment for each protein reported was determined using a custom script based on the FC-B score calculation from the reference Mellacheruvu et al., 2013. The CRAPome: a contaminant repository for affinity purification mass spectrometry data. Nat. Methods. 10(8):730-736. Doi:10.1038/nmeth.2557. The target protein, HNF1A, was identified as the 1st ranked enriched protein and the 1st ranked transcription factor based on IP-Mass Spectrometry.
- Submitted by
- Mark Mackiewicz
- Lab
- Richard Myers, HAIB
- Grant
- U54HG006998
- Download
- HNF1A_2_IP-MS.png
HNF1A (Homo sapiens)
compliant
- Caption
- HepG2 whole cell lysate was immunoprecipitated using the primary antibody (GeneTex; GTX113850). The IP fraction was loaded on a 12% Bio-Rad TGX gel and separated with the Bio-Rad Tetra Cell system. A gel fragment (rectangle outline) corresponding to the bands indicated on the Coomassie Blue stained gel image were excised and sent to the University of Alabama at Birmingham Cancer Center Mass Spectrometry/Proteomics Shared Facility. Analysis of gel fragment 3 from HepG2: The sample was analyzed on a LTQ XL Linear Ion Trap Mass Spectrometer by LC-ESI-MS/MS. Peptides were identified using SEQUEST tandem mass spectral analysis with probability based matching at p < 0.05. SEQUEST results were reported with ProteinProphet protXML Viewer (TPP v4.4 JETSTREAM) and filtered for a minimum probability of 0.9. All protein hits that met these criteria were reported, including common contaminants. Fold enrichment for each protein reported was determined using a custom script based on the FC-B score calculation from the reference Mellacheruvu et al., 2013. The CRAPome: a contaminant repository for affinity purification mass spectrometry data. Nat. Methods. 10(8):730-736. Doi:10.1038/nmeth.2557. The target protein, HNF1A, was identified as the 7th ranked enriched protein and the 1st ranked transcription factor based on IP-Mass Spectrometry.
- Submitted by
- Mark Mackiewicz
- Lab
- Richard Myers, HAIB
- Grant
- U54HG006998
- Download
- HNF1A_3_IP-MS.png
HNF1A (Homo sapiens)
HepG2
not submitted for review by lab
- Reviewer comment
- IP gel for mass spectrometry analysis
- Submitted by
- Mark Mackiewicz
- Lab
- Richard Myers, HAIB
- Grant
- U54HG006998
- Download
- HNF1A_IP-MS_Coomassie_Gel.png
HNF1A (Homo sapiens)
HepG2
compliant
- Caption
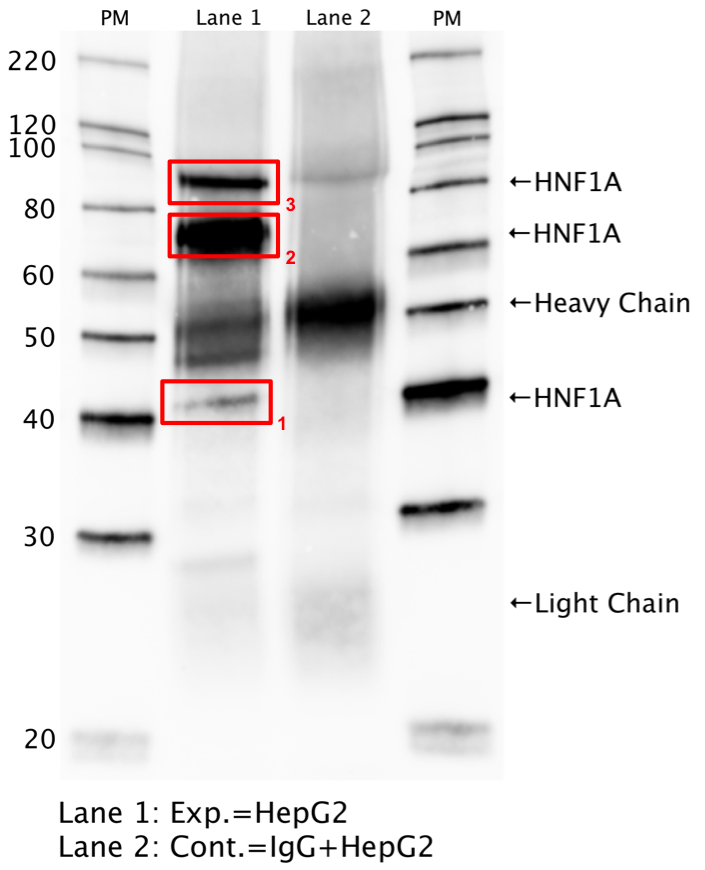
- Whole cell lysates of HepG2 were immunoprecipitated using the primary antibody (GeneTex; GTX113850). The IP fraction was separated on a 12% acrylamide gel with the Bio-Rad PROTEAN II xi system. After separation, the samples were transferred to a nitrocellulose membrane with an Invitrogen iBlot system. The membrane was probed with the primary antibody (same as that used for IP) and a secondary HRP-conjugated antibody. The resulting bands were visualized with SuperSignal West Femto Solution (Thermo Scientific). Protein Marker (PM) is labeled in kDa. Three bands were detected at ~42, 73, and 90 kDa.
- Reviewer comment
- Band 2 is closest to expected size and is about half of the overall signal in the lane
- Submitted by
- Mark Mackiewicz
- Lab
- Richard Myers, HAIB
- Grant
- U54HG006998
- Download
- HNF1A_IP-WB.png