ENCAB615WUN
Antibody against Aequorea victoria eGFP
Homo sapiens
HEK293
characterized to standards
- Status
- released
- Source (vendor)
- Abcam
- Product ID
- ab-290 (Lot GR194813-1)
- Lot ID
- GR194813-1
- Characterized targets
- eGFP (Aequorea victoria)
- Host
- rabbit
- Clonality
- polyclonal
- Purification
- antiserum
- Isotype
- IgG
- Antigen description
- Recombinant full length protein corresponding to GFP
- Aliases
- michael-snyder:AS-1667
- External resources
Characterizations
eGFP-ZSCAN26 (Homo sapiens)
Method: motif enrichment
compliant
- Caption
- The motif for target eGFP-ZSCAN26 is represented by the attached position weight matrix (PWM) derived from ENCFF957YKG. Motif enrichment analysis was done by Dr. Zhizhuo Zhang (Broad Institute, Kellis Lab) using known motifs (http://compbio.mit.edu/encode-motifs/) and previously published ChIP-seq data (http://www.broadinstitute.org/~zzhang/motifpipeline/data/TrainSetInfo.txt). The accept probability score of the given transcription factor was calculated using a Bayesian approach. This analysis also includes three motif enrichment scores, computed by overlapping the motif instances with the given ChIP-seq peak locations, as well as an enrichment rank. For more information on the underlying statistical methods, please see the attached document. Accept probability score: 0.7952650714 Global enrichment Z-score: 6.94561508003 Positional bias Z-score: 3.06652257649 Peak rank bias Z-score: 4.54167728324 Enrichment rank: 1.0
- Submitted by
- Jessika Adrian
- Lab
- Michael Snyder, Stanford
- Grant
- U54HG006996
eGFP (Aequorea victoria)
HEK293
compliant
- Caption
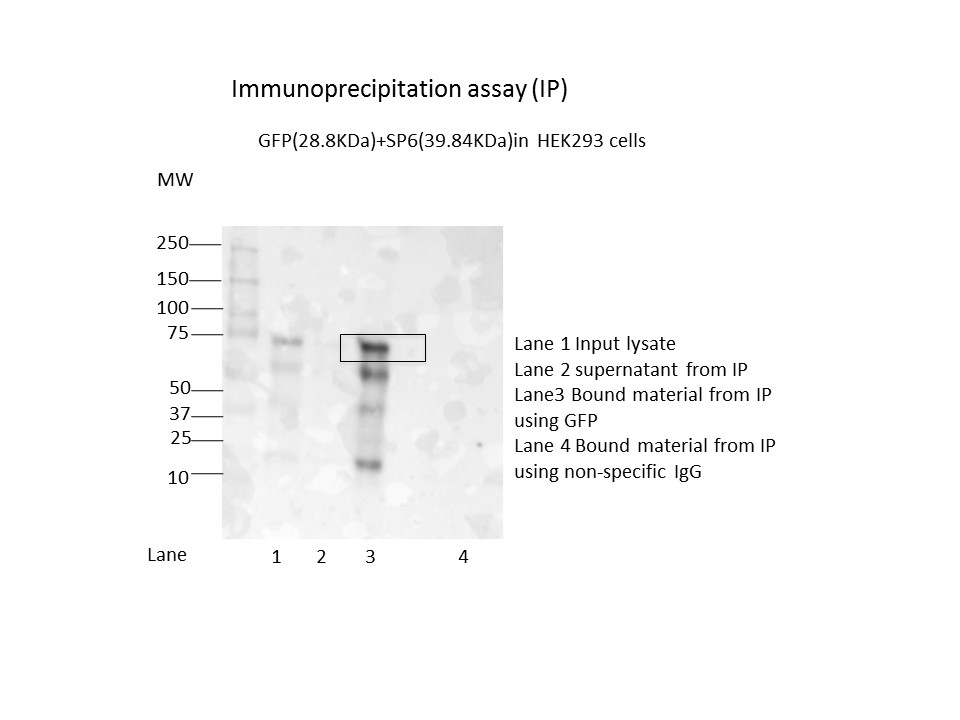
- Immunoprecipitation was performed on nuclear extracts from the cell line: HEK293 using the antibody ab-290 (Lot GR194813-1). The image shows western blot analysis of input, flowthrough, immunoprecipitate, and mock immunoprecipitate using IgG. Target molecular weight: 28.8.
- Reviewer comment
- While the western blot was passed strictly on the account that the marked signal comprises of >=50% of the total immunoreactive signal in the lane, the presence of multiple bands is not ideal. The lab will try to repeat the characterization on a different eGFP-tagged HEK293 sample that has not already been crosslinked first like this eGFP-SP6 one.
- Submitted by
- Nathaniel Watson
- Lab
- Michael Snyder, Stanford
- Grant
- U54HG006996
- Download
- GFP(GR194813-1).jpg