ENCAB463LOX
Antibody against Homo sapiens DNMT1
Homo sapiens
K562
characterized to standards with exemption
- Status
- released
- Source (vendor)
- Active Motif
- Product ID
- 39204
- Lot ID
- 08714024
- Characterized targets
- DNMT1 (Homo sapiens)
- Host
- mouse
- Clonality
- monoclonal
- Purification
- affinity
- Aliases
- michael-snyder:AS-843
- External resources
Characterizations
DNMT1 (Homo sapiens)
K562
compliant
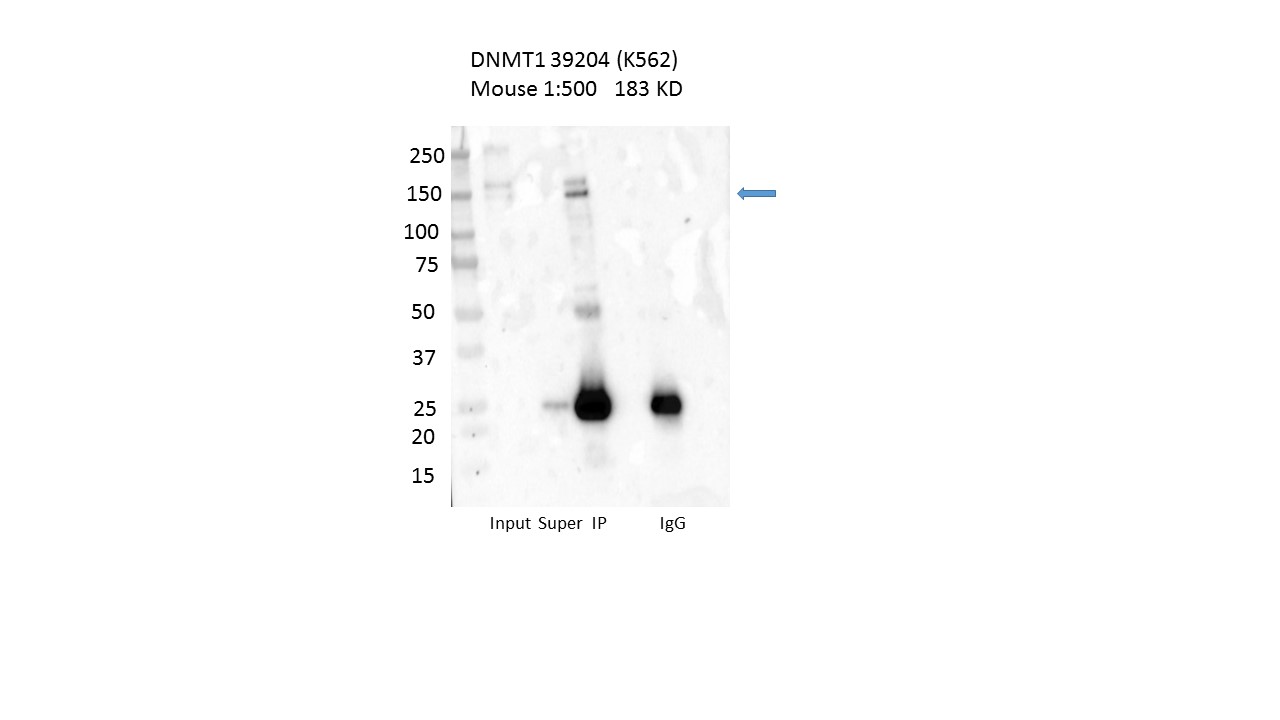
- Caption
- Immunoprecipitation was performed on nuclear extracts from the cell line: K562 using the antibody 39204. The image shows western blot analysis of input, flowthrough, immunoprecipitate, and mock immunoprecipitate using IgG. Target molecular weight: 183.165.
- Submitter comment
- According to UniProt (P26358), isoform 1 (P26358-1) is ~183 kD and isoform 3 (P26358-3) is ~144 kD.
- Submitted by
- Nathaniel Watson
- Lab
- Michael Snyder, Stanford
- Grant
- U54HG006996
- Download
- #1143 K562 DNMT1(39204).jpg
DNMT1 (Homo sapiens)
K562
not compliant
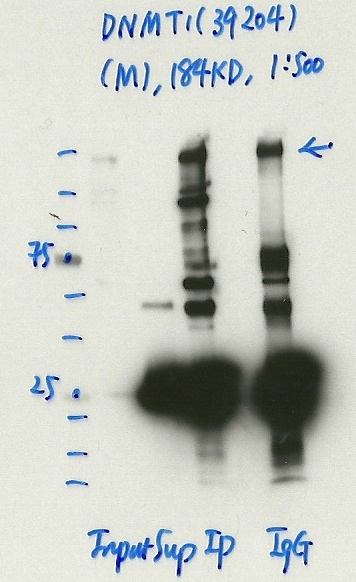
- Caption
- Immunoprecipitation was performed on nuclear extracts from the cell line: K562, using the antibody 39204. The blot shows western blot analysis of input, flowthrough, immunoprecipitate and mock immunoprecipitate using IgG.
- Reviewer comment
- Indicated band not enriched over IgG. Also difficult to discern if at appropriate size relative to expected.
- Submitted by
- Denis Salins
- Lab
- Michael Snyder, Stanford
- Grant
- U54HG006996
DNMT1 (Homo sapiens)
K562
compliant
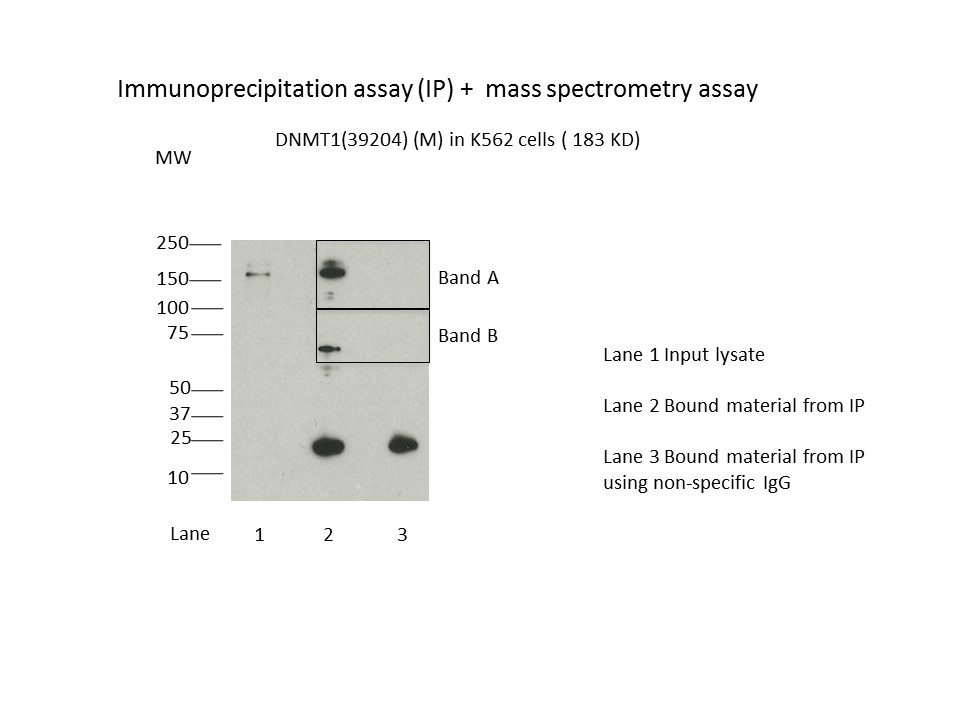
- Caption
- Immunoprecipitation was performed on nuclear extracts from the cell line K562 using the antibody 39204. Lane 1: input nuclear lysate. Lane 2: material immunoprecipitated with antibody. Lane 3: material immunoprecipitated using control IgG. Marked bands were excised from gel and subjected to analysis by mass spectrometry. Target molecular weight: 183.165.
- Submitted by
- Nathaniel Watson
- Lab
- Michael Snyder, Stanford
- Grant
- U54HG006996
- Download
- DNMT1_39204-1.jpg
DNMT1 (Homo sapiens)
Method: immunoprecipitation followed by mass spectrometry
exempt from standards
- Caption
- IP followed by mass spectrometry. Briefly, protein was immunoprecipitated from K562 nuclear cell lysates using the antibody 39204, and the IP fraction was loaded on a 10% polyacrylamide gel (NuPAGEBis-Tris Gel) and separated with an Invitrogen NuPAGE electrophoresis system. The gel was stained by ColloidialCoomassie G-250 stain, gel fragments corresponding to the bands indicated were excised. Then proteins were trypsinized using the in-gel digestion method. Digested proteins were analyzed on an Orbitrap Elite mass spectrometer (Thermo Scientific) by the nanoLC-ESI-MS/MS technique. Peptides were identified by the SEQUEST algorithm and filtered with a high confidence threshold (Peptide false discovery rate < 1%, 2 unique peptides per protein minimum, mass error < 10 ppm).
- Submitter comment
- CCAR1 were detected in both Band A and Band B. Both CCAR1 and DNMT1 have interactions with TP53 (44KD) and SIRT7 (45KD), http://thebiogrid.org/108123/summary/homo-sapiens/dnmt1.html and http://thebiogrid.org/120867/summary/homo-sapiens/ccar1.html. TP53 and SIRT7 are not in the same gel with CCAR1 and DNMT1. Both EIF3A (Band A) and EIF3D (Band B) involves poly(A) RNA binding and are translation initiation factor. Both DIDO1 (244KD) (Band A) and DNMT1 have interactions with EED (50KD), http://thebiogrid.org/108123/summary/homo-sapiens/dnmt1.html and http://thebiogrid.org/116266/summary/homo-sapiens/dido1.html, EED is not in the same gel slice with DIDO1 and DNMT1. Both NCOA5 (Band B) and DNMT1 have interactions with B9D2 (19KD), http://thebiogrid.org/108123/summary/homo-sapiens/dnmt1.html and http://thebiogrid.org/121747/summary/homo-sapiens/ncoa5.html, B9D2 is not in the same gel slice with NCOA5 and DNMT1 (band B). Both PUF60 (Band B) and DNMT1 have interactions with CDK2 (34KD), http://thebiogrid.org/116502/summary/homo-sapiens/puf60.html and http://thebiogrid.org/108123/summary/homo-sapiens/dnmt1.html, but CDK2 is not in the same gel slice with DNMT1 and PUF60 (band B).
- Reviewer comment
- Although DIDO1 which is a putative TF is ranked higher than DNMT1 in band A, the submitters have noted that they have common interacting partners.
- Submitted by
- Nathaniel Watson
- Lab
- Michael Snyder, Stanford
- Grant
- U54HG006996
- Download
- DNMT1_39204_final.pdf