ENCAB053JZL
Antibody against Homo sapiens GSPT2
Homo sapiens
at least one cell type or tissue, K562
not characterized to standards
- Status
- released
- Source (vendor)
- MBLI
- Product ID
- RN119PW
- Lot ID
- 001
- Characterized targets
- GSPT2 (Homo sapiens)
- Host
- rabbit
- Clonality
- polyclonal
- External resources
Characterizations
GSPT2 (Homo sapiens)
not compliant
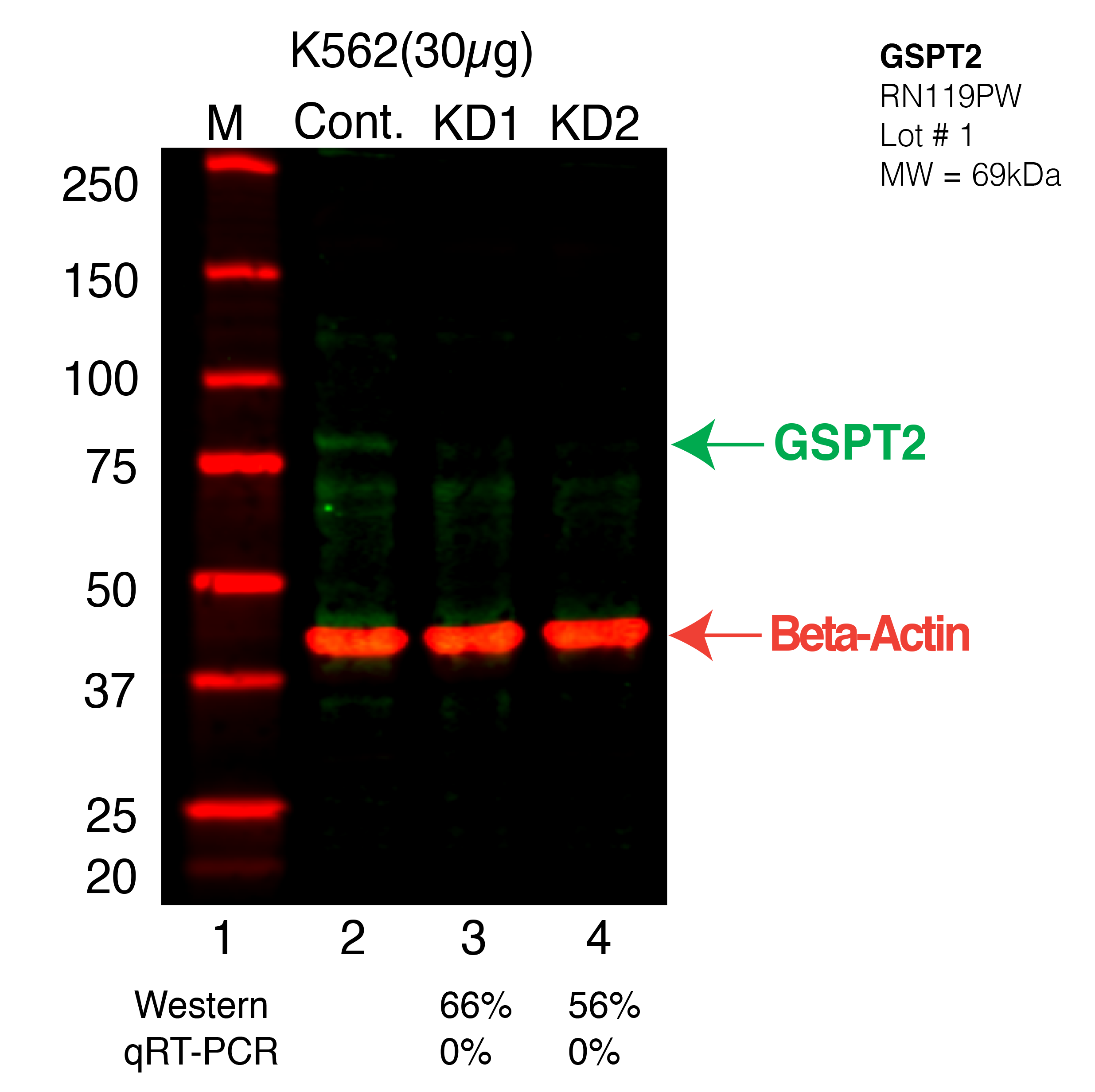
- Caption
- Western blot following CRISPR against GSPT2 in K562 whole cell lysate using GSPT2 specific antibody. Lane 1 is a ladder, lane 2 is K562 non-targeting control knockdown, lane 3 and 4 are two different CRISPR against GSPT2. GSPT2 protein appears as the green arrow, Beta-actin serves as a control and appears in red arrow.
- Submitted by
- Xintao Wei
- Lab
- Brenton Graveley, UConn
- Grant
- U41HG009889
- Download
- GSPT2-K562-CRISPR-RN119PW.png
GSPT2 (Homo sapiens)
K562
not compliant
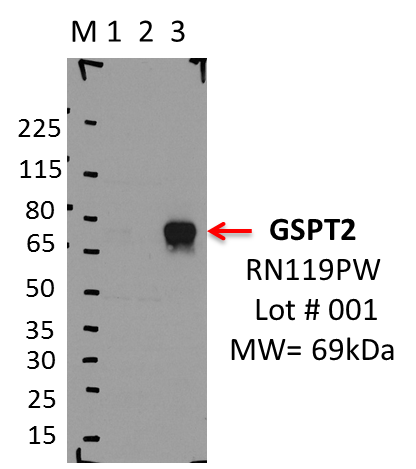
- Caption
- IP-WB analysis of K562 whole cell lysate using the GSPT2 specific antibody, RN119PW. Lane 1 is 2.5% of five million whole cell lysate input. Lanes 2 and 3 are 50% of IP enrichment from five million whole cell lysate using normal IgG antibody and the GSPT2-specific antibody, RN119PW. The same antibody was used to detect protein levels via Western blot. This antibody passes preliminary validation and will be further pursued for secondary validation. *NOTE* Protein sizes are taken from Genecards.org and are only estimates based on sequence. Actual protein size may differ based on protein characteristics and electrophoresis method used.
- Reviewer comment
- Lane changed to 3 (2022/08/08).
- Submitted by
- Steven Blue
- Lab
- Gene Yeo, UCSD
- Grant
- U41HG009889
- Download
- MBL_RN119PW_001_GSPT2.png
GSPT2 (Homo sapiens)
not submitted for review by lab
- Caption
- IP-WB analysis of K562 whole cell lysate using GSPT2 specific antibody. Lane 1 is 2.5% of five million whole cell lysate Input, lane 2 is 2.5% of supernatant after immunoprecipitation and Lane 3 is 50% of IP enrichment using rabbit polyclonal Anti-GSPT2 (eRF3b) (Human) pAb. This antibody passes preliminary validation and will be further pursued for primary and secondary validation.
- Submitted by
- Balaji Sundararaman
- Lab
- Gene Yeo, UCSD
- Grant
- U54HG007005
- Download
- MBLI_RN119PW_001_GSPT2.png