ENCAB001RIV
Antibody against Homo sapiens IRF2
Homo sapiens
K562
characterized to standards
- Status
- released
- Source (vendor)
- Santa Cruz Biotech
- Product ID
- sc-13042
- Lot ID
- I1603
- Characterized targets
- IRF2 (Homo sapiens)
- Host
- rabbit
- Clonality
- polyclonal
- Aliases
- michael-snyder:IRF2-human_sc-13042_I1603
- External resources
Characterizations
IRF2 (Homo sapiens)
Method: motif enrichment
compliant
- Caption
- The motif for target IRF2 is represented by the attached position weight matrix (PWM) derived from ENCFF284BAX. Motif enrichment analysis was done by Dr. Zhizhuo Zhang (Broad Institute, Kellis Lab). Accept probability score: 0.658136505228 Global enrichment Z-score: 7.31182165197 Positional bias Z-score: 11.7100005936 Peak rank bias Z-score: 10.4205916377 Enrichment rank: 1.0
- Submitted by
- Kathrina Onate
- Lab
- Michael Snyder, Stanford
- Grant
- U54HG006996
IRF2 (Homo sapiens)
K562
compliant
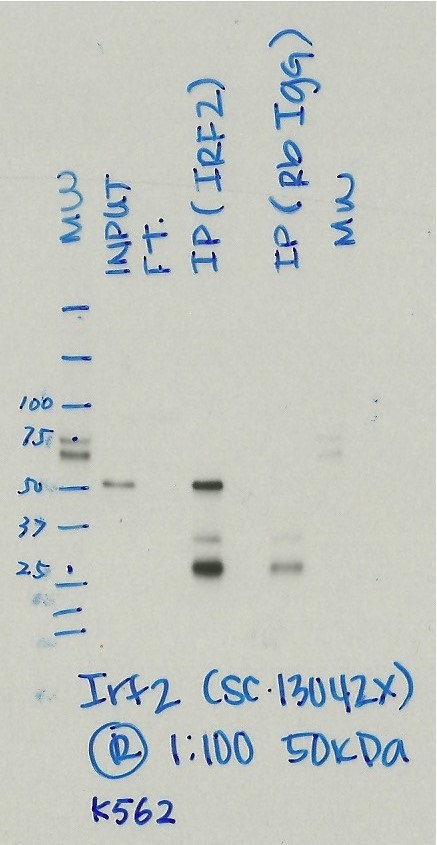
- Caption
- Immunoprecipitation was performed on nuclear extracts from the cell line: K562, using the antibody sc-13042. The blot shows western blot analysis of input, flowthrough, immunoprecipitate and mock immunoprecipitate using IgG.Molecular Weight: 39.354
- Submitted by
- Nathaniel Watson
- Lab
- Michael Snyder, Stanford
- Grant
- U54HG006996
IRF2 (Homo sapiens)
Method: immunoprecipitation followed by mass spectrometry
compliant
- Caption
- IP followed by mass spectrometry: Briefly, protein was immunoprecipitated from K562 whole cell lysates using sc-13042, and the IP fraction was loaded on a 10% polyacrylamide gel (NuPAGE Bis-Tris Gel) and separated with an Invitrogen NuPAGE electrophoresis system. The gel was silver-stained, gel fragments corresponding to the bands indicated were excised and destained using the SilverSNAP Stain for Mass Spectrometry (Pierce). Then proteins were treated with trypsin using the in-gel digestion method. Digested proteins were analyzed on a LTQ-Orbitrap (Thermo Scientific) b the nanoLC-ESI-MS/MS technique. Peptides were identified by the SEQUEST algorithm and filtered with a high confidence threshold (protein false discovery rate <1%, 2 peptides per protein minimum).
- Reviewer comment
- There are only 4 peptides from distinct proteins detected and all are equally ranked...
- Submitted by
- Kathrina Onate
- Lab
- Michael Snyder, Stanford
- Grant
- U54HG004558
- Download
- IRF2_final Sheet1.pdf
IRF2 (Homo sapiens)
K562
compliant
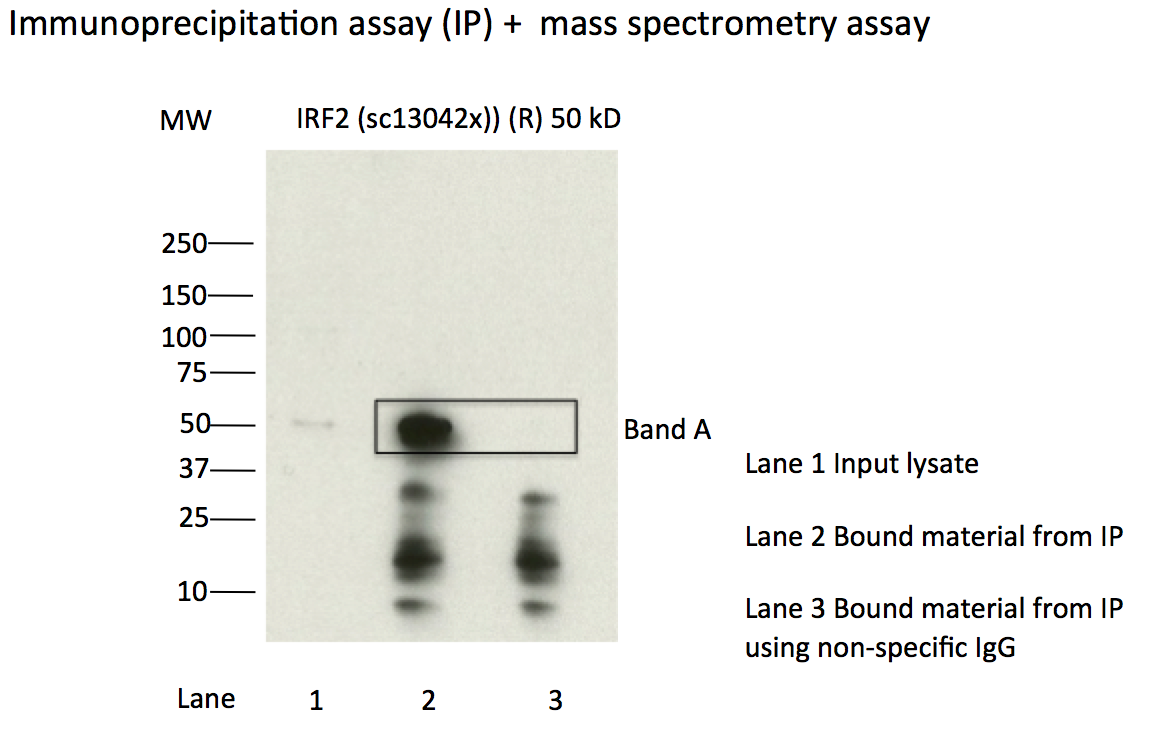
- Caption
- Immunoprecipitation of IRF2 from K562 cells using sc-13042. Lane 1: input nuclear lysate. Lane 2: material immunoprecipitated with sc-13042. Lane 3: material immunoprecipitated using control IgG. Band A was excised from gel and subject to analysis by mass spectrometry. Expected band size ~50kD.
- Submitted by
- Kathrina Onate
- Lab
- Michael Snyder, Stanford
- Grant
- U54HG004558
- Download
- IRF2_IP.png