ENCAB000BDV
Antibody against Homo sapiens BMI1
Homo sapiens
GM12878, K562, HeLa-S3, HepG2, IMR-90, A549, heart, endothelial cell of umbilical vein, MCF-7, HEK293T
characterized to standards
Homo sapiens
H1
not characterized to standards
- Status
- released
- Source (vendor)
- Cell Signaling
- Product ID
- 6964S
- Lot ID
- 1
- Characterized targets
- BMI1 (Homo sapiens)
- Host
- rabbit
- Clonality
- monoclonal
- Purification
- affinity
- Isotype
- IgG
- Antigen description
- The carboxy terminus of human Bmi1 protein.
- Aliases
- bradley-bernstein:PchAb 774
- External resources
Characterizations
BMI1 (Homo sapiens)
HEK293T
compliant
- Caption
- Immunoprecipitation was performed on nuclear extracts from the cell line: HEK293T using the antibody 6964S. The image shows western blot analysis of input, flowthrough, immunoprecipitate, and mock immunoprecipitate using IgG. Target molecular weight: 41, 43.
- Submitted by
- Nathaniel Watson
- Lab
- Michael Snyder, Stanford
- Grant
- U54HG006996
- Download
- #1144 HEK293T Bmi1(6964S).jpg
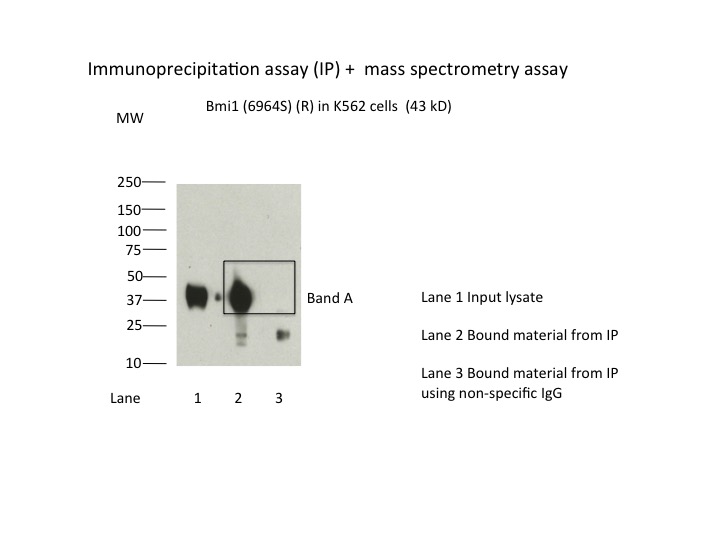
BMI1 (Homo sapiens)
K562
compliant
- Caption
- Immunoprecipitation of Bmi1 from K562 cells using 6964S. Lane 1: input nuclear lysate. Lane 2: material immunoprecipitated with 6964S. Lane 3: material immunoprecipitated using control IgG. Band A was excised from gel and subject to analysis by mass spectrometry. Expected band size is 43 kDa.
- Submitted by
- Kathrina Onate
- Lab
- Michael Snyder, Stanford
- Grant
- U54HG006996
- Download
- Bmi1.jpg
BMI1 (Homo sapiens)
Method: immunoprecipitation followed by mass spectrometry
compliant
- Caption
- IP followed by mass spectrometry: Briefly, protein was immunoprecipitated from K562 nuclear cell lysates using 6964S, and the IP fraction was loaded on a 10% polyacrylamide gel (NuPAGEBis-Tris Gel) and separated with an Invitrogen NuPAGE electrophoresis system. The gel was stained by ColloidialCoomassie G-250 stain, gel fragments corresponding to the bands indicated were excised. Then proteins were trypsinized using the in-gel digestion method. Digested proteins were analyzed on an Orbitrap Elite mass spectrometer (Thermo Scientific) by the nanoLC-ESI-MS/MS technique. Peptides were identified by the SEQUEST algorithm and filtered with a high confidence threshold (Peptide false discovery rate < 1%, 2 unique peptides per protein minimum, mass error < 10 ppm).
- Submitter comment
- For this factor, we found DNA-binding proteins SMARCE1, DPF2, RING1, RNF2 and ACTL6A have more peptides than targeted protein Bmi1. We would like to explain as follows: Bmi1 has unique interaction with DPF2, and DPF2 also has interaction with SMARCE1, which one can find here: http://thebiogrid.org/111909/summary/homo-sapiens/dpf2.html SMARCE1 has interactions with RING1, RNF2 and ACTL6A, it can be found here: http://thebiogrid.org/112489/summary/homo-sapiens/smarce1.html
- Submitted by
- Kathrina Onate
- Lab
- Michael Snyder, Stanford
- Grant
- U54HG006996
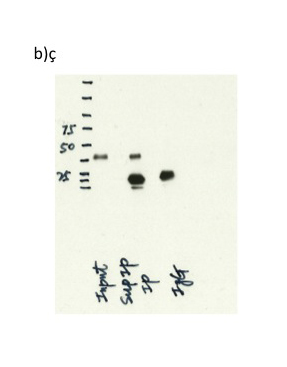
BMI1 (Homo sapiens)
GM12878K562HeLa-S3HepG2IMR-90A549heart
compliant
- Caption
- Western blot analysis of nuclear lysates prepared from multiple cells lines loaded in the order : GM12878, K562, HelaS3, HepG2, IMR90, A549 and Heart using the antibody 6964S. Molecular mass: 36949 Da
- Submitted by
- Trupti Kawli
- Lab
- Michael Snyder, Stanford
- Grant
- U54HG006996
- Download
- BMI1_6964S_WB_a.jpg
BMI1 (Homo sapiens)
K562
compliant
- Caption
- b) Immunoprecipitation was performed on nuclear extracts from the cell line: K562, using the antibody 6964S .The blot shows western blot analysis of input, flowthrough, immunoprecipitate and mock immunoprecipitate using IgG. Expected size ~37 kDa.
- Submitted by
- Trupti Kawli
- Lab
- Michael Snyder, Stanford
- Grant
- U54HG006996
- Download
- BMI1_6964S_WB_b.jpg
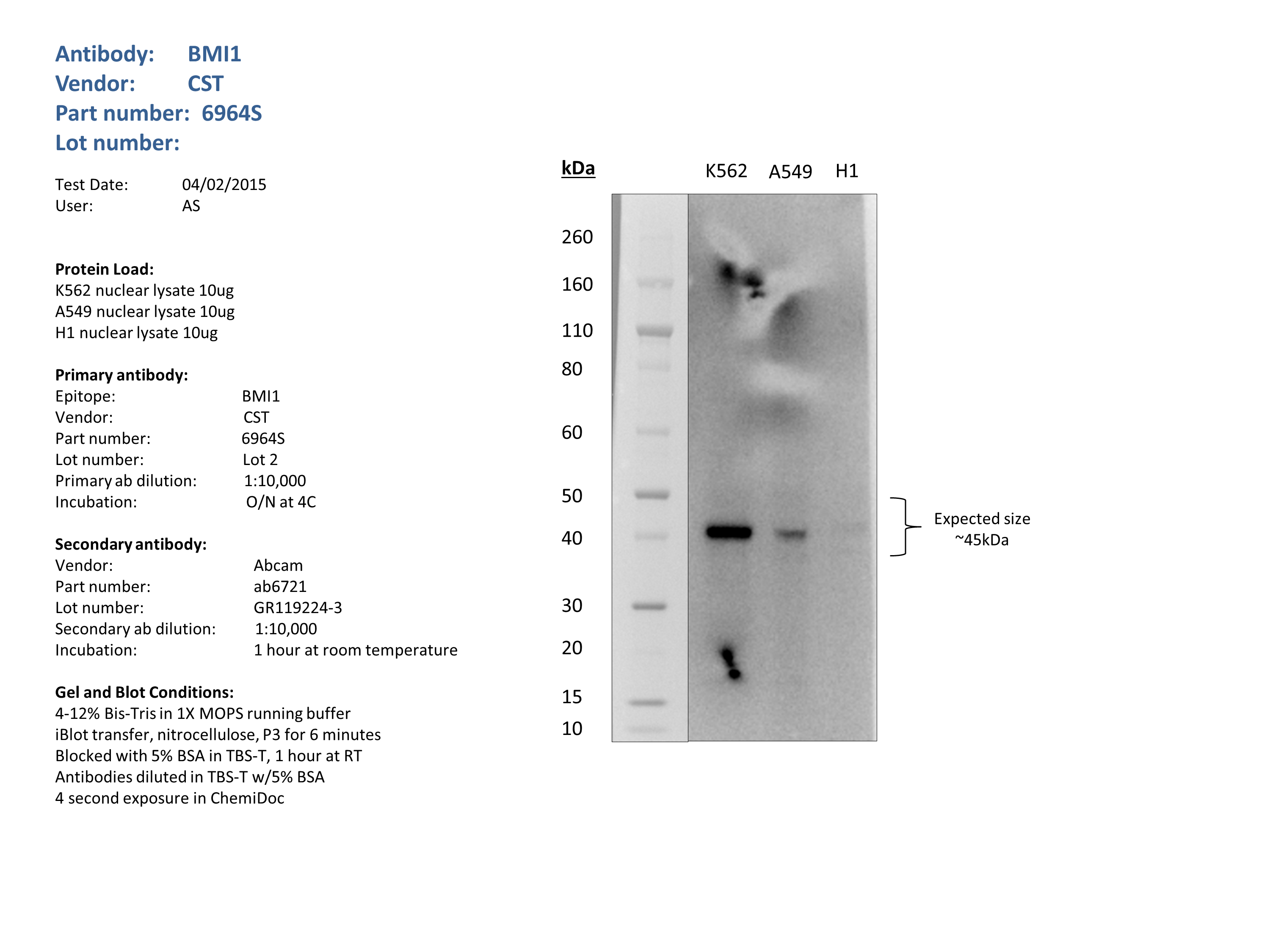
BMI1 (Homo sapiens)
K562A549H1
compliant
- Caption
- Nuclear lysates from K562 (10ug), A549 (10ug) and H1 (10ug) were loaded into a 4-12% Bis-Tris gel in 1X MOPS running buffer. After separation, the samples were transferred to a nitrocellulose membrane using iblot. Membrane was blocked for an hour in room temperature, with 5% BSA in TBS-T and blotted with primary antibody in the appropriate concentration over night at 4c. Membrane was washed and blotted with secondary HRP-conjugated antibody. Detection was made with Optiblot ECL Detect Kit (ab133406) for 2 min. Only one band at the expected size (45kDa) was detected.
- Submitted by
- Noam Shoresh
- Lab
- Bradley Bernstein, Broad
- Grant
- U54HG006991
- Download
- BMI1_CST_6964S_Lot2_V2.png
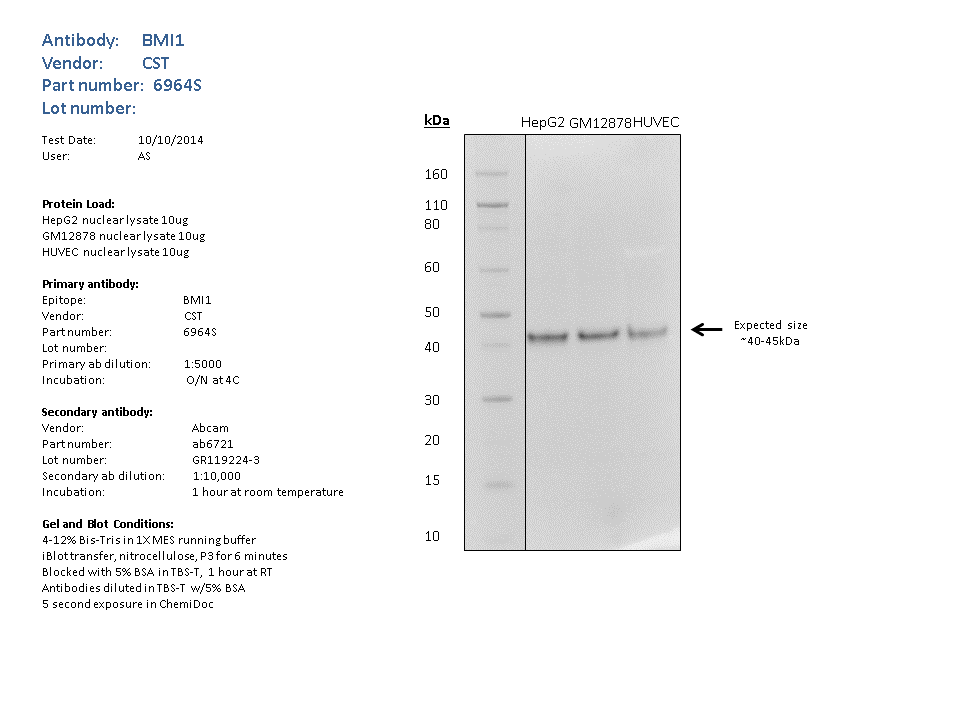
BMI1 (Homo sapiens)
HepG2GM12878endothelial cell of umbilical vein
compliant
- Caption
- Nuclear lysates from GM12878 (10ug),HUVEC (10ug), HepG2 (10ug), were loaded into a 4-12% Bis-Tris gel in 1X MES running buffer. After separation, the samples were transferred to a nitrocellulose membrane using iblot. Membrane was blocked for an hour in room temperature, with 5% BSA in TBS-T and blotted with primary antibody in the appropriate concentration over night at 4c. Membrane was washed and blotted with secondary HRP-conjugated antibody. Detection was made with Optiblot ECL Detect Kit (ab133406) for 2 min. The only band detected was of the expected size (~40kDa). **Molecular mass: 36949 Da**
- Submitted by
- Noam Shoresh
- Lab
- Bradley Bernstein, Broad
- Grant
- U54HG006991
- Download
- BMI1_CST_6964S_WB.png
BMI1 (Homo sapiens)
MCF-7
compliant
- Caption
- Immunoprecipitation was performed on nuclear extracts from the cell line: MCF-7, using the antibody 6964S. The blot shows western blot analysis of input, flowthrough, immunoprecipitate and mock immunoprecipitate using IgG.
- Submitted by
- Denis Salins
- Lab
- Michael Snyder, Stanford
- Grant
- U54HG006996
- Download
- expt1032_4.jpg