ENCAB000ALF
Antibody against Homo sapiens STAT1
Homo sapiens
HeLa-S3
characterized to standards
Homo sapiens
GM23338, GM12878
characterized to standards with exemption
Homo sapiens
any cell type or tissue
partially characterized
- Status
- released
- Source (vendor)
- Santa Cruz Biotech
- Product ID
- sc-345
- Lot ID
- G2406
- Characterized targets
- STAT1 (Homo sapiens)
- Lot ID aliases
- D2908, K2106
- Host
- rabbit
- Clonality
- polyclonal
- Purification
- affinity
- Antigen description
- STAT1 (alpha) p-91
- External resources
Characterizations
STAT1 (Homo sapiens)
GM12878
exempt from standards
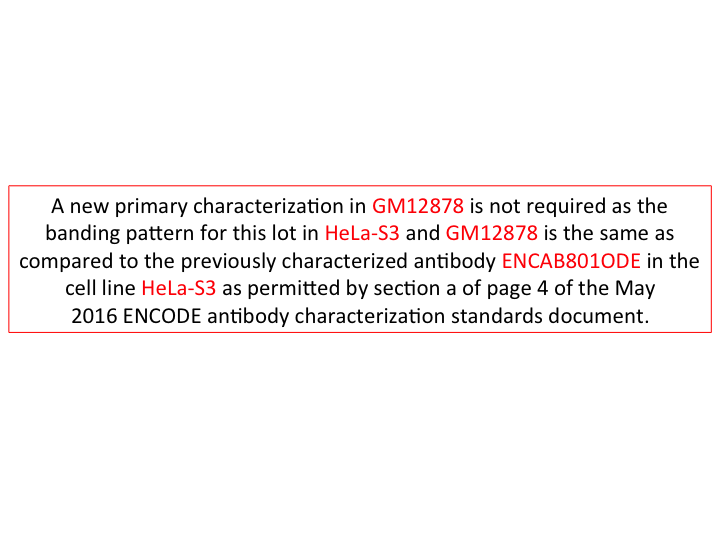
- Caption
- A new primary characterization for GM12878 should not be required as the banding pattern for characterized cell lines in common (HeLa-S3) with another fully characterized lot ENCAB801ODE of the same antibody (sc-345). Primary characterization of ENCAB801ODE also exhibits the same banding pattern.
- Submitter comment
- We think this antibody lot should be exempted from primary characterization in GM12878 as we've demonstrated that the lot behaves the same in immunoprecipitations as another lot (ENCAB801ODE) in GM12878 and HeLa-S3.
- Reviewer comment
- This antibody was exempted from primary characterization in GM12878 by the ENCODE antibody review panel on October 17, 2016.
- Submitted by
- Jessika Adrian
- Lab
- Michael Snyder, Stanford
- Grant
- U54HG006996
- Download
- ENCAB00ALF_primary_GM12878.png
STAT1 (Homo sapiens)
Method: motif enrichment
compliant
- Caption
- The motif for target STAT1 is represented by the attached position weight matrix (PWM) derived from ENCFF760TJN. Motif enrichment analysis was done by Dr. Zhizhuo Zhang (Broad Institute, Kellis Lab). Accept probability score: 0.728533116472 Global enrichment Z-score: 3.57768729177 Positional bias Z-score: 1.97817636299 Peak rank bias Z-score: 1.31732457347 Enrichment rank: 7.0
- Submitted by
- Kathrina Onate
- Lab
- Michael Snyder, Stanford
- Grant
- U54HG006996
STAT1 (Homo sapiens)
Method: immunoblot
not reviewed
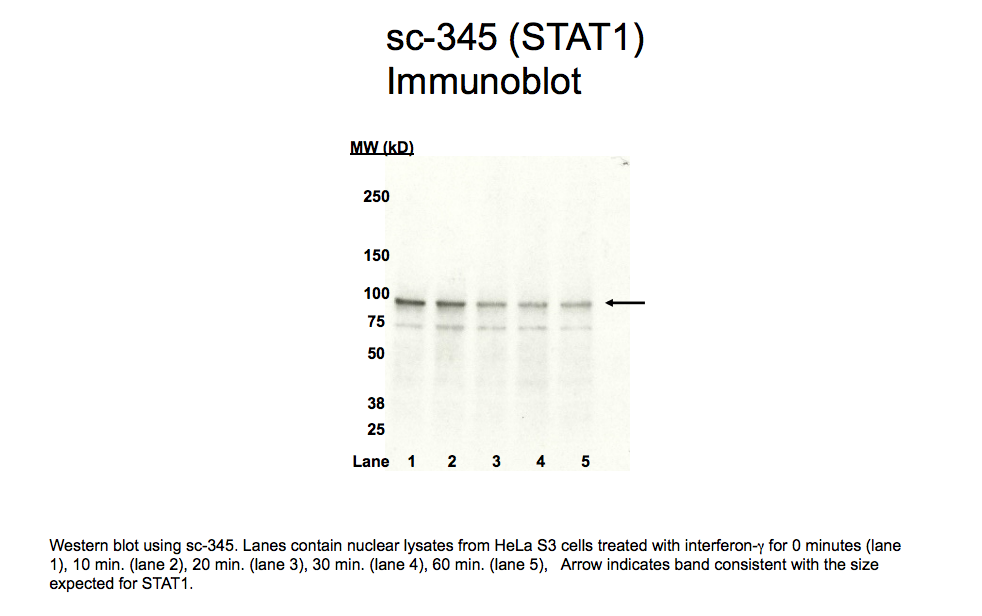
- Caption
- A predominant band consistent with the size expected for STAT1 (~91kD, indicated by arrow) is observed in HeLa S3 lysates using sc-345. Therefore, sc-345 meets this criterion for validation. Western blot using sc-345. Lanes contain nuclear lysates from HeLa S3 cells treated with interferon-γ for 0 minutes (lane 1), 10 min. (lane 2), 20 min. (lane 3), 30 min. (lane 4), 60 min. (lane 5), Arrow indicates band consistent with the size expected for STAT1.
- Submitted by
- Michael Snyder
- Lab
- Michael Snyder, Stanford
- Grant
- U54HG004558
- Download
- human_STAT1_validation_Snyder.pdf
STAT1 (Homo sapiens)
Method: motif enrichment
not reviewed
- Caption
- Calculations were done by Pouya Kheradpour using a collection of known motifs available at http://www.broadinstitute.org/%7Epouyak/motif-disc/human/. Table 1 shows the fold-enrichments, enrichment p-values and fraction of peaks which contain the motif. The motif which produced the largest value for each criterion is shown in Table 1. Note that while the maximally enriched motifs may differ from the motif with the highest enrichment p-values and the most represented motif, the motifs are highly similar (Figure 1) and thus all values are similar between motifs. Motifs were identified using a matching stringency corresponding to 4-6 (6-mer). Peaks identified by IDR (1% cutoff) were used in the analysis and +/-50bp from peak centers were considered. Enrichments are for a given motif vs. a background consisting of +/- 50bp from the centers of all DnaseI hypersensitive peaks. Repeat mask/simple repeats from UCSC and all gencode v7 exons (including non-protein coding genes) were excluded from the analysis. Comparison to shuffle motifs were used to correct for compositional bias. Enrichment is the corrected # of motifs in ChIP peaks/corrected # of motifs in DNaseI peaks. The current ENCODE standard calls for >4-fold enrichment and >10% motif representation for this criteria to be used for validation. The 5 STAT1 and STAT2 datasets presented here exceed these thresholds and the antibody is considered validated. In addition, we note that this antibody is also validated based on the overlap between data obtained with antibody sc-345 (STAT1) and antibody sc-476 (STAT2), which are known to form a complex upon interferon- stimulation. Details can be found in the validation document for sc-476.
- Submitted by
- Michael Snyder
- Lab
- Michael Snyder, Stanford
- Grant
- U54HG004558
- Download
- human_STAT1_validation_Snyder.pdf
STAT1 (Homo sapiens)
GM23338
exempt from standards
- Caption
- The ENCODE Binding Working Group finds for some valuable biosamples that recreating a primary on well characterized antibodies is not cost effective. Therefore, they allow exemption from standards for these samples.
- Submitter comment
- The lab is asking for an exemption forIPSCs due to the lack of resources to make a primary characterization for them
- Reviewer comment
- Exempted by the Feb 29, 2016 antibody review panel
- Submitted by
- Richard Myers
- Lab
- Richard Myers, HAIB
- Grant
- U54HG006998
- Download
- No_biosample.png
STAT1 (Homo sapiens)
HeLa-S3
compliant
- Caption
- A predominant band consistent with the size expected for STAT1 (~91kD, indicated by arrow) is observed in HeLa S3 lysates using sc-345. Therefore, sc-345 meets this criterion for validation. Western blot using sc-345. Lanes contain nuclear lysates from HeLa S3 cells treated with interferon-γ for 0 minutes (lane 1), 10 min. (lane 2), 20 min. (lane 3), 30 min. (lane 4), 60 min. (lane 5), Arrow indicates band consistent with the size expected for STAT1.
- Submitted by
- Kathrina Onate
- Lab
- Michael Snyder, Stanford
- Grant
- U54HG004558
- Download
- WB Snyder ALF.png