ENCAB000AKJ
Antibody against Homo sapiens RFX5
Homo sapiens
HeLa-S3, K562, GM12878
characterized to standards
Homo sapiens
MCF-7, A549
characterized to standards with exemption
Homo sapiens
any cell type or tissue
partially characterized
Homo sapiens
HepG2
not characterized to standards
- Status
- released
- Source (vendor)
- Rockland
- Product ID
- 200-401-194
- Lot ID
- 14562
- Characterized targets
- RFX5 (Homo sapiens)
- Host
- rabbit
- Clonality
- polyclonal
- Antigen description
- Raised against RFX5 peptide corresponding to a region at amino acids 320 to 494 of the human protein conjugated to Keyhole Limpet Hemocyanin.
- External resources
Characterizations
RFX5 (Homo sapiens)
MCF-7A549
exempt from standards
- Caption
- No more lot 14562 was available to characterize it in MCF-7 and A549 cell lines. Instead, we characterized a different lot of the same antibody in those cell lines as well as in K562 to use for comparison purposes to demonstrate that the lots behave similarly.
- Submitter comment
- We would like an exemption for this lot under a variation of Guideline b listed on page 4 of the May 2016 TF Antibody Standards document.
- Reviewer comment
- An exemption was granted by the ENCODE Antibody Review Panel on July 27, 2017
- Submitted by
- Jessika Adrian
- Lab
- Michael Snyder, Stanford
- Grant
- U54HG006996
- Download
- ENCAB000AKJ_A549_MCF7_primary.png
RFX5 (Homo sapiens)
Method: motif enrichment
compliant
- Caption
- The motif for target RFX5 is represented by the attached position weight matrix (PWM) derived from ENCFF745KVW. Motif enrichment analysis was done by Dr. Zhizhuo Zhang (Broad Institute, Kellis Lab). Accept probability score: 0.868714667886 Global enrichment Z-score: 5.32133003348 Positional bias Z-score: 6.77380255701 Peak rank bias Z-score: 3.84165801819 Enrichment rank: 3.0
- Submitted by
- Kathrina Onate
- Lab
- Michael Snyder, Stanford
- Grant
- U54HG006996
RFX5 (Homo sapiens)
Method: motif enrichment
not reviewed
- Caption
- Calculations were done by Pouya Kheradpour using a collection of known motifs (9 motifs were considered) available at http://www.broadinstitute.org/%7Epouyak/motif-disc/human/. Table 1 shows the fold-enrichments, enrichment p-values and fraction of peaks which contain the motif. The motif which produced the largest value for each criterion is shown in Table 1. Note that while the maximally enriched motif differs from the motif with the highest enrichment p-values and the most represented motif, the motifs are highly similar (Figure 1) and thus all values are similar between motifs. Motifs were identified using a matching stringency corresponding to 4-6 (6-mer). Peaks identified by IDR (1% cutoff) were used in the analysis and +/-50bp from peak centers were considered. Enrichments are for a given motif vs. a background consisting of +/- 50bp from the centers of all DnaseI hypersensitive peaks. Repeat mask/simple repeats from UCSC and all gencode v7 exons (including non-protein coding genes) were excluded from the analysis. Comparison to shuffle motifs were used to correct for compositional bias. Enrichment is the corrected # of motifs in ChIP peaks/corrected # of motifs in DNaseI peaks. The current ENCODE standard calls for >4-fold enrichment and >10% motif representation for this criteria to be used for validation. As shown in Table 1, the ENCODE datasets for RFX5 in GM12878, H1 hES, and HepG2 cells exceed these thresholds and the antibody is considered validated.
- Submitted by
- Michael Snyder
- Lab
- Michael Snyder, Stanford
- Grant
- U54HG004558
- Download
- human_RFX5_validation_Snyder.pdf
RFX5 (Homo sapiens)
Method: immunoblot
not reviewed
- Caption
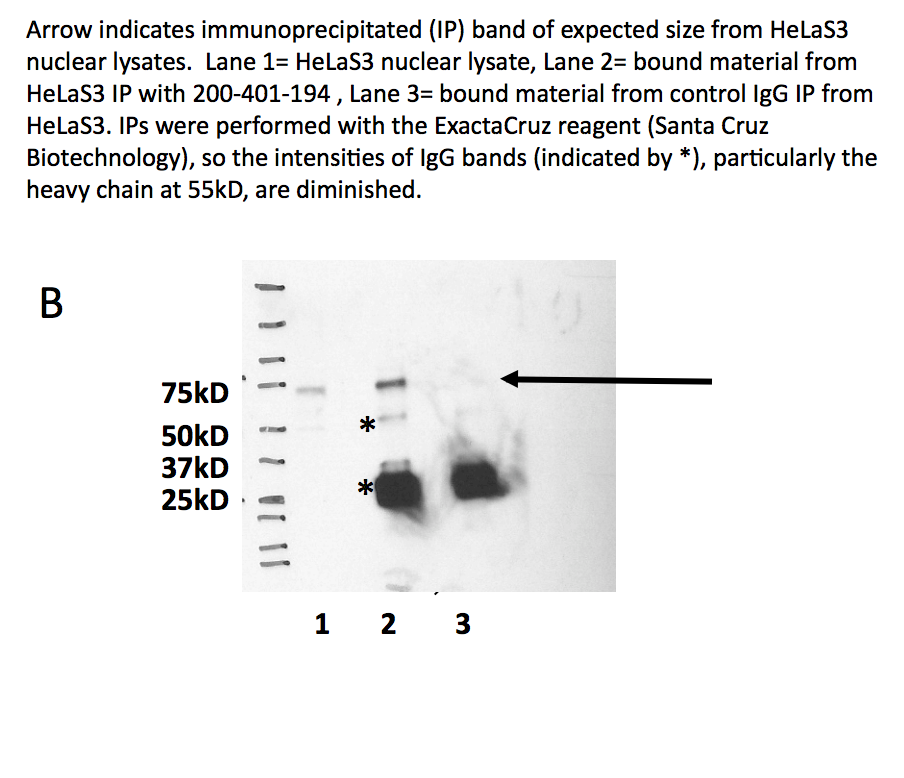
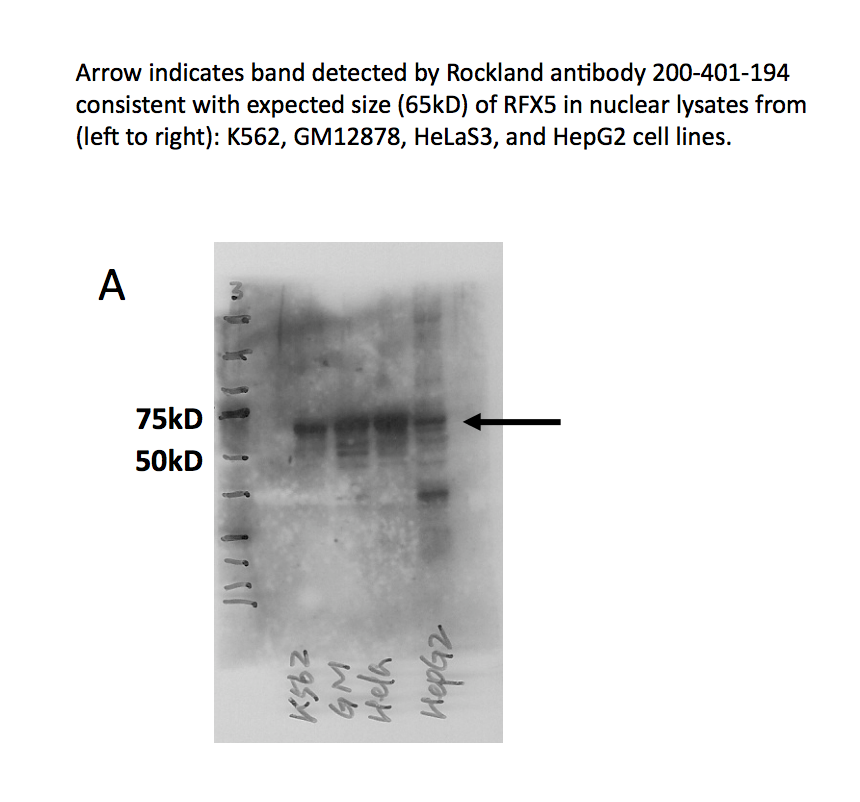
- Panel A demonstrates that a major band of ~65kD is observed in all cell lines tested. A band at ~35kD is also observed in HepG2 cells- we are unsure of the identity of this band but it may be a degradation product. Panel B demonstrates that this band is efficiently immunoprecipitated from HeLa nuclear extracts. Based on these observations, this antibody meets this ENCODE criterion.
- Submitted by
- Michael Snyder
- Lab
- Michael Snyder, Stanford
- Grant
- U54HG004558
- Download
- human_RFX5_validation_Snyder.pdf
RFX5 (Homo sapiens)
HeLa-S3
compliant
- Caption
- Panel B demonstrates that this band is efficiently immunoprecipitated from HeLa nuclear extracts. Based on these observations, this antibody meets this ENCODE criterion. Arrow indicates immunoprecipitated (IP) band of expected size fro HeLa-S3 nuclear lysates. Lane 1 = HeLa-S3 nuclear lysate. Lane 2 = bound material from HeLa-S3 IP with 200-401-194. Lane 3 = bound material from control IgG IP from HeLa-S3. IPs were performed with ExactaCruz reagent (Santa Cruz Biotechnology), so the intensities of IgG bands (indicated by *), particularly the heavy chain at 55 kD, are diminished.
- Submitted by
- Kathrina Onate
- Lab
- Michael Snyder, Stanford
- Grant
- U54HG004558
- Download
- IP Snyder AKJ.png
RFX5 (Homo sapiens)
K562
compliant
- Caption
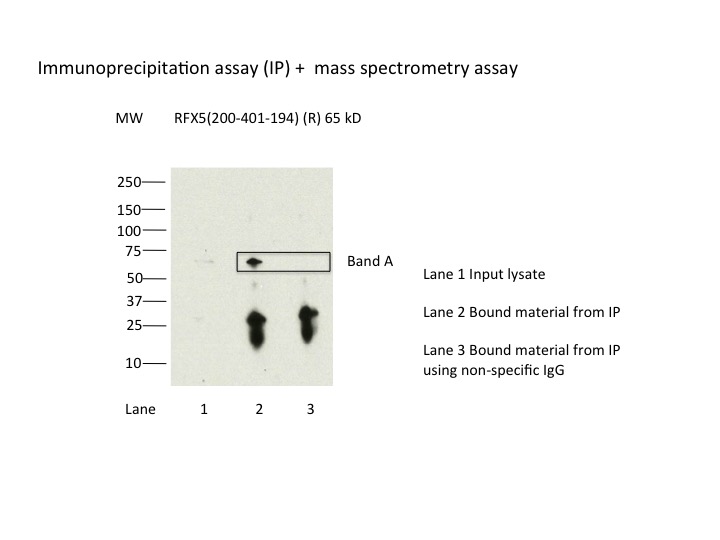
- Immunoprecipitation was performed on nuclear extracts from the cell line K562 using the antibody 200-401-194. Lane 1: input nuclear lysate. Lane 2: material immunoprecipitated with antibody. Lane 3: material immunoprecipitated using control IgG. Marked bands were excised from gel and subjected to analysis by mass spectrometry. Target molecular weight: 65.3.
- Submitted by
- Nathaniel Watson
- Lab
- Michael Snyder, Stanford
- Grant
- U54HG006996
RFX5 (Homo sapiens)
Method: immunoprecipitation followed by mass spectrometry
compliant
- Caption
- IP followed by mass spectrometry. Briefly, protein was immunoprecipitated from K562 nuclear cell lysates using the antibody 200-401-194, and the IP fraction was loaded on a 10% polyacrylamide gel (NuPAGEBis-Tris Gel) and separated with an Invitrogen NuPAGE electrophoresis system. The gel was stained by ColloidialCoomassie G-250 stain, gel fragments corresponding to the bands indicated were excised. Then proteins were trypsinized using the in-gel digestion method. Digested proteins were analyzed on an Orbitrap Elite mass spectrometer (Thermo Scientific) by the nanoLC-ESI-MS/MS technique. Peptides were identified by the SEQUEST algorithm and filtered with a high confidence threshold (Peptide false discovery rate < 1%, 2 unique peptides per protein minimum, mass error < 10 ppm).
- Submitted by
- Nathaniel Watson
- Lab
- Michael Snyder, Stanford
- Grant
- U54HG006996
- Download
- RFX5_final.pdf
RFX5 (Homo sapiens)
K562GM12878HeLa-S3HepG2
compliant
- Caption
- Panel A demonstrates that a major band of ~65kD is observed in all cell lines tested. A band at ~35kD is also observed in HepG2 cells- we are unsure of the identity of this band but it may be a degradation product. Arrow indicates band detected by Rockland antibody 200-401-194 consistent with expected size (65kD) of RFX5 in nuclear lysates from (left to right): K562, GM12878, HeLa-S3, and HepG2 cell lines.
- Reviewer comment
- Lane 4 not compliant because band is not 50% of total signal
- Submitted by
- Kathrina Onate
- Lab
- Michael Snyder, Stanford
- Grant
- U54HG004558
- Download
- WB Snyder AKJ.png