ENCAB000AHY
Antibody against Homo sapiens IRF3
Homo sapiens
HepG2, GM12878, K562, SK-N-SH
characterized to standards
Homo sapiens
any cell type or tissue
partially characterized
- Status
- released
- Source (vendor)
- Santa Cruz Biotech
- Product ID
- sc-9082
- Lot ID
- I0908
- Characterized targets
- IRF3 (Homo sapiens)
- Host
- rabbit
- Clonality
- polyclonal
- Antigen description
- Epitope corresponding to amino acids 1-425 of IRF-3 of human origin.
- External resources
Characterizations
IRF3 (Homo sapiens)
Method: immunoblot
not reviewed
- Submitted by
- Michael Snyder
- Lab
- Michael Snyder, Stanford
- Grant
- U54HG004558
- Download
- human_IRF3_validation_Snyder.pdf
IRF3 (Homo sapiens)
K562
compliant
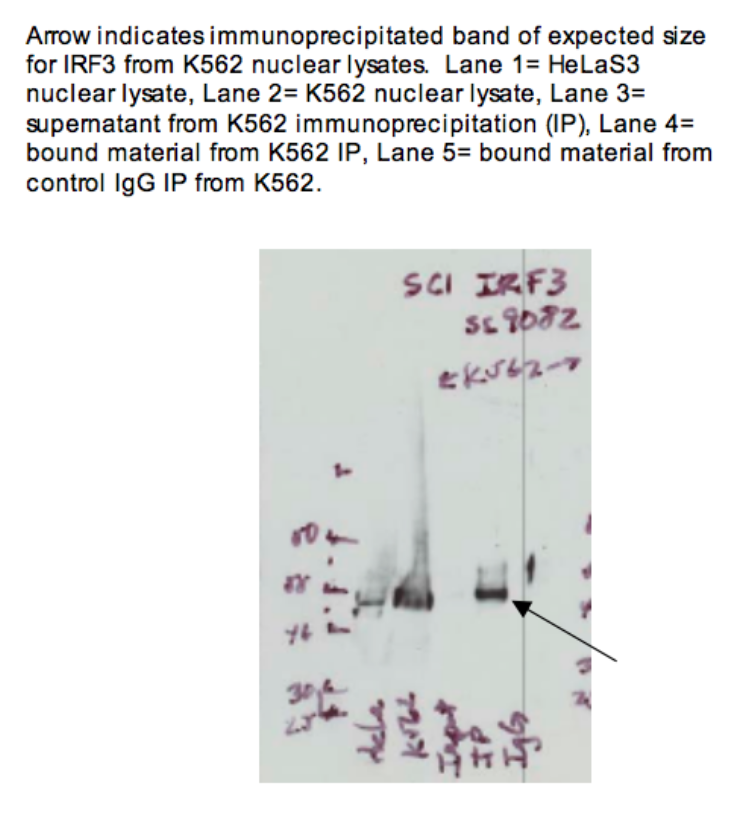
- Caption
- Arrow indicates immunoprecipitated band of expected size for IRF3 from K562 nuclear lysates. Lane 1 = HeLa-S3 nuclear lysate. Lane 2 = K562 nuclear lysate. Lane 3 = supernatant from K562 immunoprecipitation (IP) Lane 4 = bound material from K562 IP Lane 5 = bound material from control IgG IP from K562
- Submitted by
- Kathrina Onate
- Lab
- Michael Snyder, Stanford
- Grant
- U54HG004558
- Download
- IP Snyder AHY.png
IRF3 (Homo sapiens)
SK-N-SH
compliant
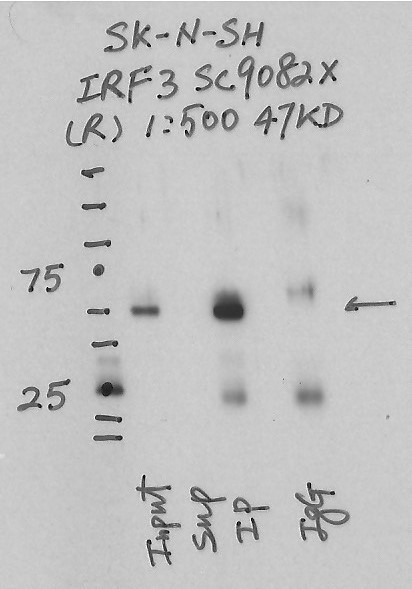
- Caption
- Immunoprecipitation was performed on nuclear extracts from the cell line: SK-N-SH using the antibody sc-9082. The image shows western blot analysis of input, flowthrough, immunoprecipitate, and mock immunoprecipitate using IgG. Target molecular weight: 47KD.
- Submitted by
- Nathaniel Watson
- Lab
- Michael Snyder, Stanford
- Grant
- U54HG006996
- Download
- IP1148 SK-N-SH IRF3 sc9082x.jpg
IRF3 (Homo sapiens)
Method: immunoprecipitation followed by mass spectrometry
compliant
- Caption
- IP followed by mass spectrometry: Briefly, protein was immunoprecipitated from K562 whole cell lysates using sc-9082, and the IP fraction was loaded on a 10% polyacrylamide gel (NuPAGE Bis-Tris Gel) and separated with an Invitrogen NuPAGE electrophoresis system. The gel was silver-stained, gel fragments corresponding to the bands indicated were excised and destained using the SilverSNAP Stain for Mass Spectrometry (Pierce). Then proteins were treated with trypsin using the in-gel digestion method. Digested proteins were analyzed on a LTQ-Orbitrap (Thermo Scientific) b the nanoLC-ESI-MS/MS technique. Peptides were identified by the SEQUEST algorithm and filtered with a high confidence threshold (protein false discovery rate <1%, 2 peptides per protein minimum).
- Submitted by
- Kathrina Onate
- Lab
- Michael Snyder, Stanford
- Grant
- U54HG004558
- Download
- IRF3_final_AHY Sheet1.pdf
IRF3 (Homo sapiens)
K562
compliant
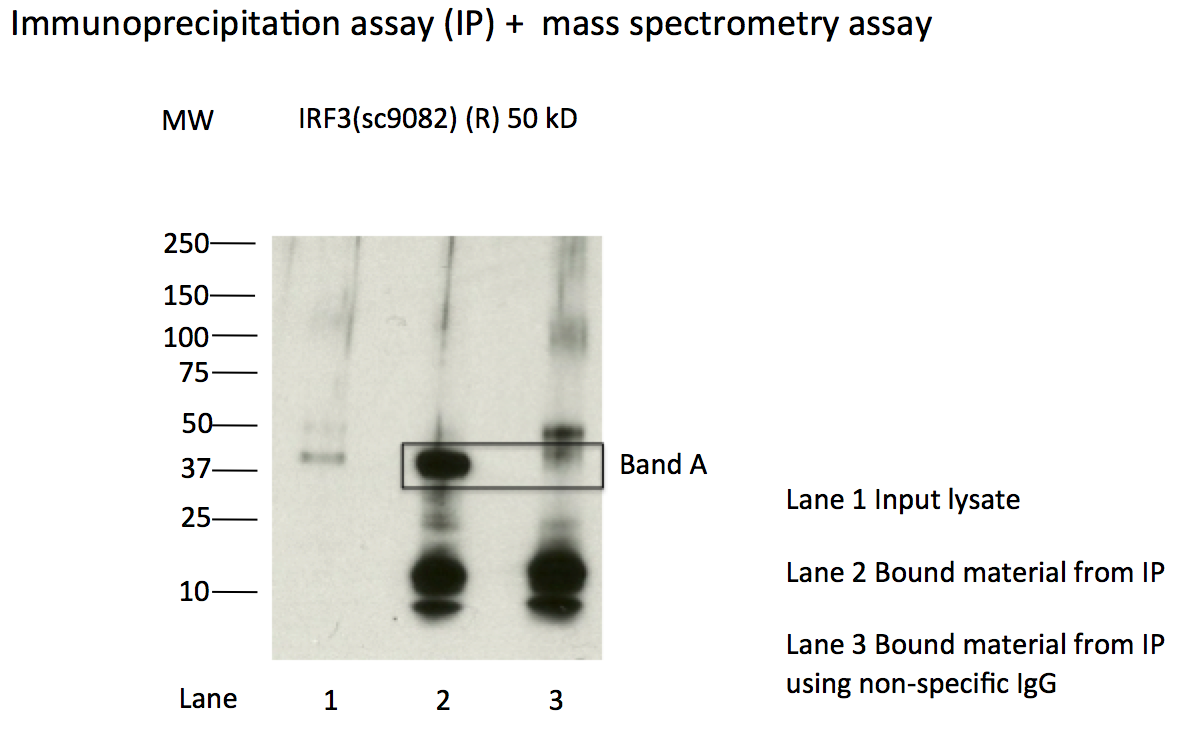
- Caption
- Immunoprecipitation of IRF3 from K562 cells using sc-9082. Lane 1: input nuclear lysate. Lane 2: material immunoprecipitated with sc-9082. Lane 3: material immunoprecipitated using control IgG. Band A was excised from gel and subject to analysis by mass spectrometry. Expected band size ~47kD.
- Submitted by
- Kathrina Onate
- Lab
- Michael Snyder, Stanford
- Grant
- U54HG004558
- Download
- IRF3_IP.png
IRF3 (Homo sapiens)
HepG2GM12878K562
compliant
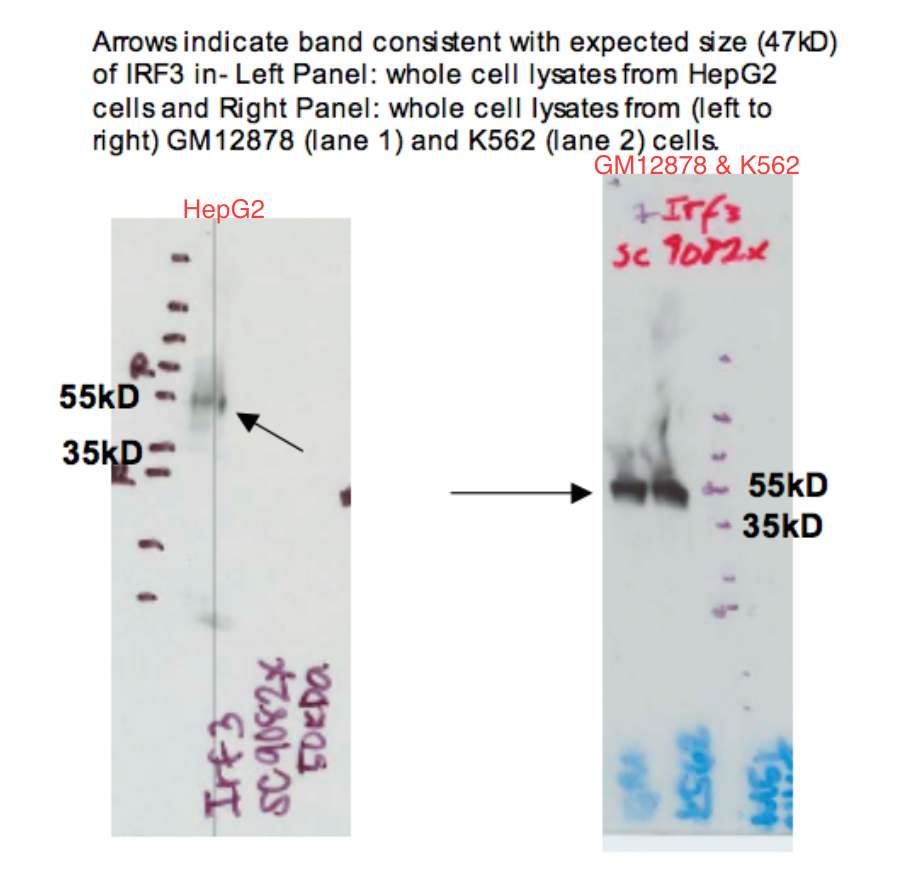
- Caption
- Arrows indicate band consistent with expected size (47kD) of IRF3 in LEFT PANEL: whole cell lysates from HepG2 cells and RIGHT PANEL: whole cell lysates of GM12878 and K562 cells (from left to right). Although there are two images for the immunoblot, we are considering them as one image. We consider HepG2 as lane 1, GM12878 as lane 2, and K562 as lane 3.
- Reviewer comment
- Although there are two images for the immunoblot, we are considering them as one image. We consider HepG2 as lane 1, GM12878 as lane 2, and K562 as lane 3.
- Submitted by
- Kathrina Onate
- Lab
- Michael Snyder, Stanford
- Grant
- U54HG004558
- Download
- WB Snyder AHY.png