ENCAB000AHI
Antibody against Homo sapiens HDAC2
Homo sapiens
K562, HepG2, GM12878, SK-N-SH, A549, H1
characterized to standards
Homo sapiens
any cell type or tissue
partially characterized
- Status
- released
- Source (vendor)
- Bethyl Labs
- Product ID
- A300-705A
- Lot ID
- 1
- Characterized targets
- HDAC2 (Homo sapiens)
- Host
- rabbit
- Clonality
- polyclonal
- Purification
- affinity
- Aliases
- bradley-bernstein:PchAb 317
- External resources
Characterizations
HDAC2 (Homo sapiens)
Method: ChIP-seq comparison
compliant
- Caption
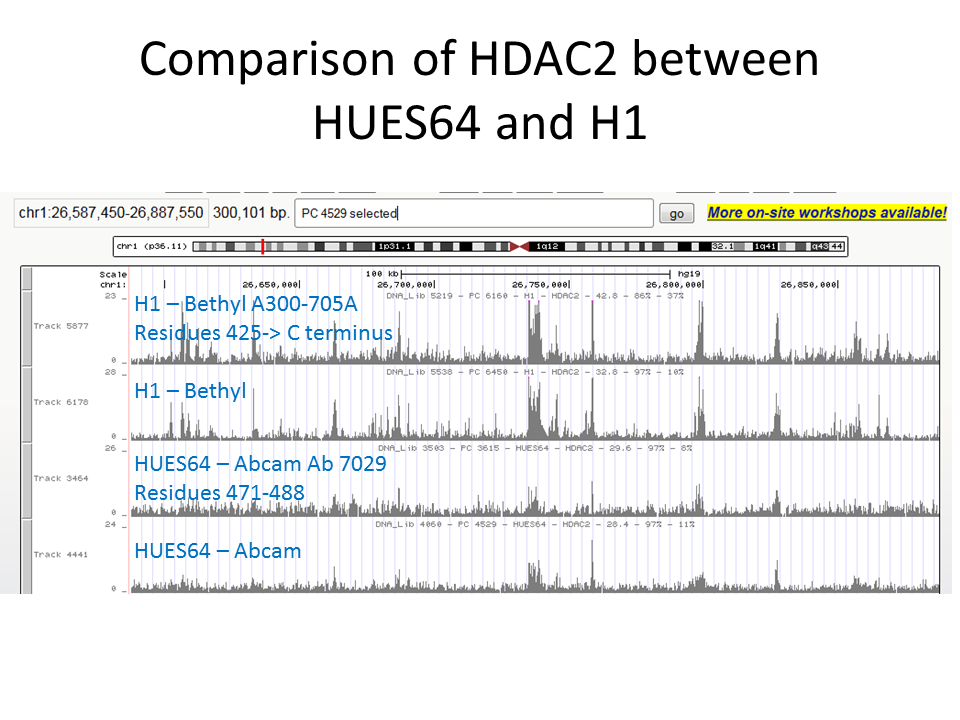
- This validation relies on the use of antibodies to different members of a known complex, in HepG2 cells, and the demonstration that highly similar patterns of enrichment are obtained with each antibody. The first track illustrates the antibody being validated (HDAC2, PchAb 317), the second track illustrates an antibody to CHD4 (Broad alias PchAb 1222), and the third track illustrates an antibody to KDM1A (Broad alias PchAb 158). Thus, this illustrates the high similarity between three members of the NuRD complex. Overall correlation scores: HDAC2 vs. CHD4: 0.8539; HDAC2 vs. KDM1A: 0.8566
- Submitted by
- Kathrina Onate
- Lab
- Bradley Bernstein, Broad
- Grant
- U54HG006991
- Download
- HDAC2 Bethyl A300-705A lot 1 SAV.pdf
HDAC2 (Homo sapiens)
not compliant
- Caption
- ChIP-seq was performed using two closely related cell types, and using different antibodies that were raised against different regions of the same protein, and highly similar results were obtained. This is suggestive that the enrichment obtained using ChIP is due to a specific interaction between the antibody and the intended cellular protein.
- Reviewer comment
- Failed due to additional information needed (needs to be compared to an eligible antibody lot, needs to list ENCAB of lot, ENCSR of data used in comparison, IDR metrics need to be reported)
- Submitted by
- Noam Shoresh
- Lab
- Bradley Bernstein, Broad
- Grant
- U54HG006991
HDAC2 (Homo sapiens)
A549H1
compliant
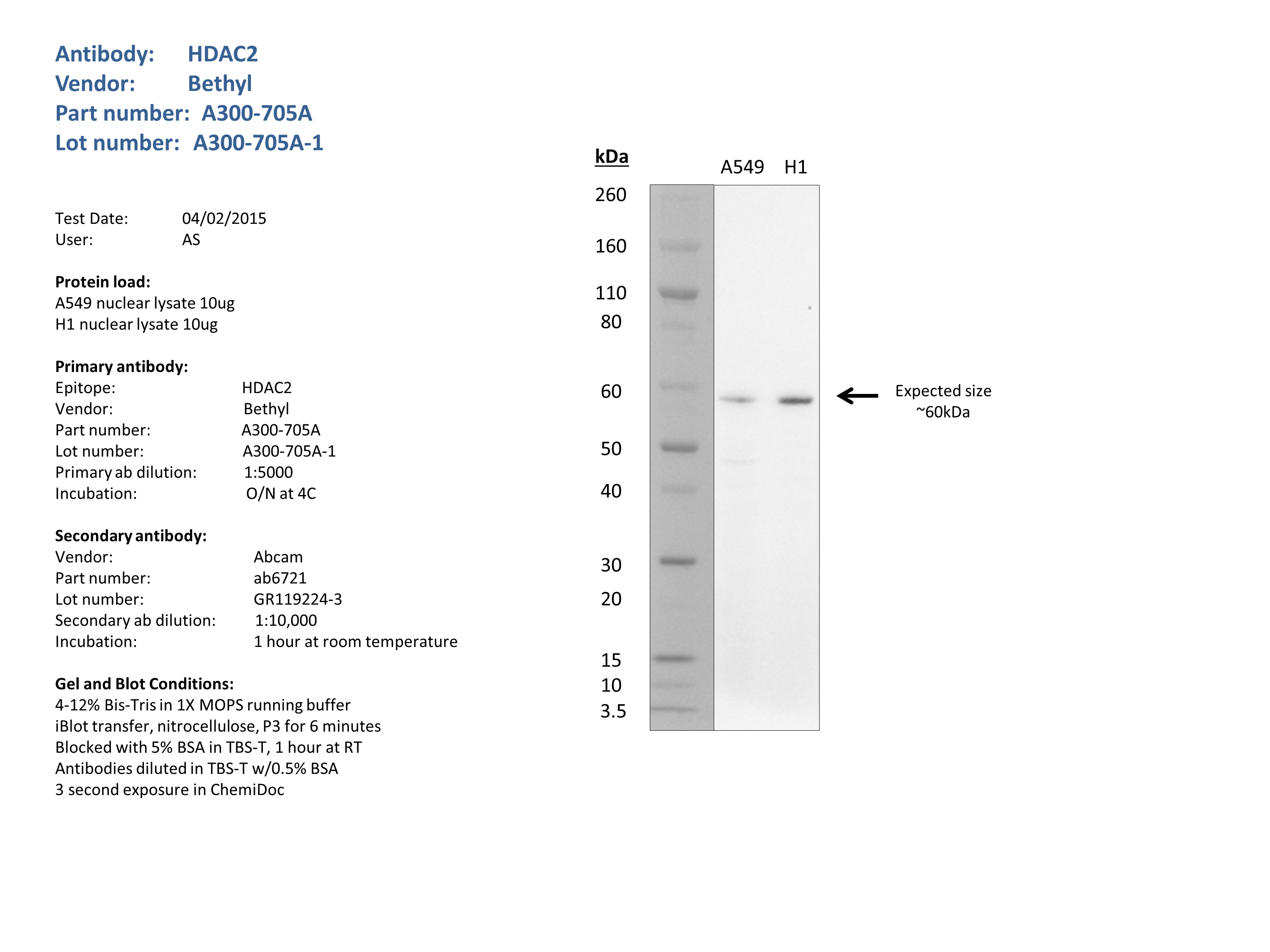
- Caption
- Nuclear lysates from A549 (10ug) and H1 (10ug) were loaded into a 4-12% Bis-Tris gel in 1X MOPS running buffer. After separation, the samples were transferred to a nitrocellulose membrane using iblot. Membrane was blocked for an hour in room temperature, with 5% BSA in TBS-T and blotted with primary antibody in the appropriate concentration over night at 4c. Membrane was washed and blotted with secondary HRP-conjugated antibody. Detection was made with Optiblot ECL Detect Kit (ab133406) for 2 min. Bands were detected at the expected size (60kDa).
- Submitted by
- Noam Shoresh
- Lab
- Bradley Bernstein, Broad
- Grant
- U54HG006991
HDAC2 (Homo sapiens)
K562HepG2GM12878SK-N-SH
compliant
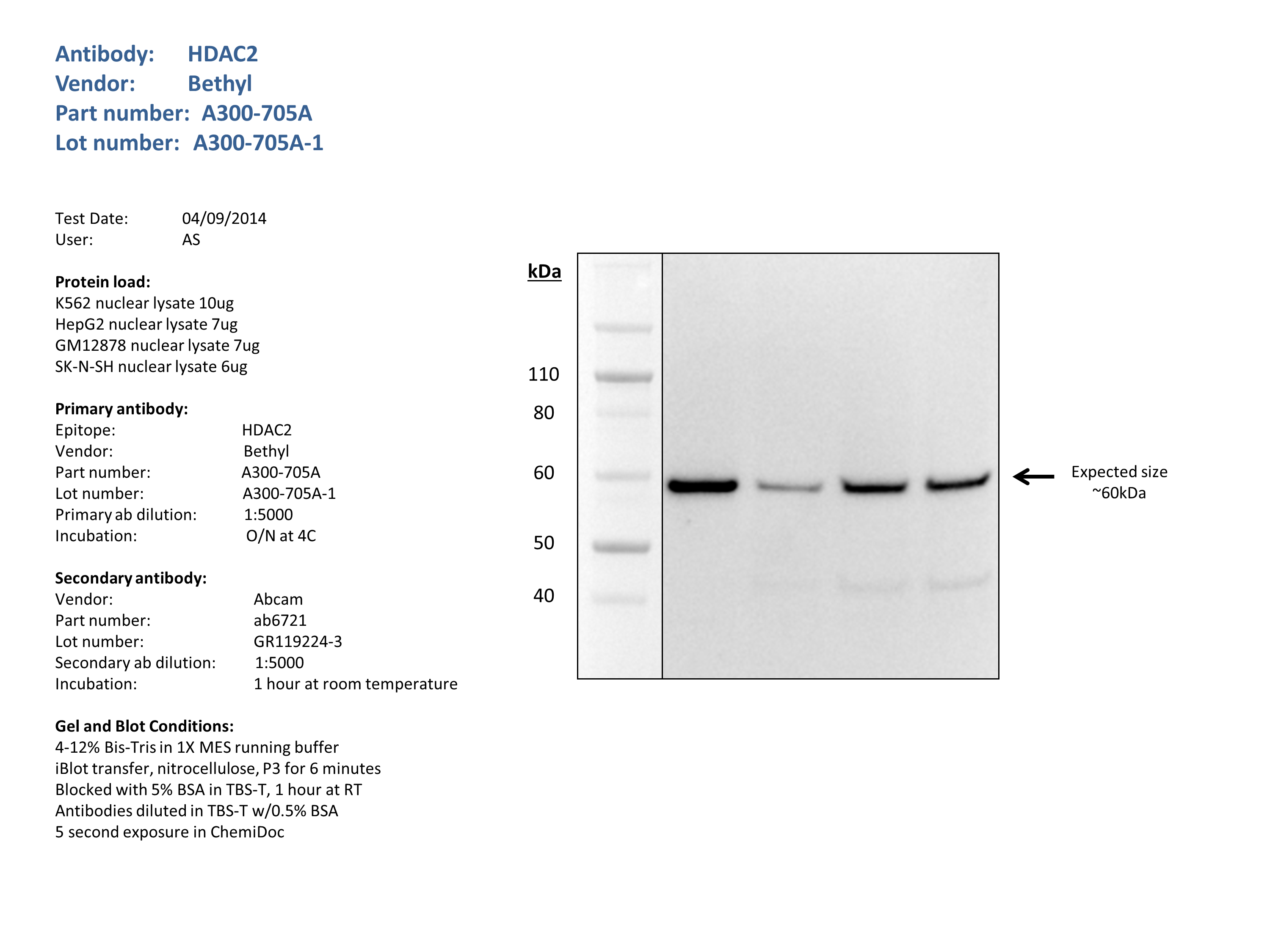
- Caption
- Nuclear lysates from K562 (10ug), HepG2 (7ug), GM12878 (7ug), SK-N-SH (6ug), were loaded into a 4-12% Bis-Tris gel in 1X MES running buffer. After separation, the samples were transferred to a nitrocellulose membrane using the iblot system. Membrane was blocked for an hour in room temperature, with 5% BSA in TBS-T and blotted with primary antibody in the appropriate concentration over night at 4c. Membrane was washed and blotted with secondary HRP-conjugated antibody. Detection was made with Optiblot ECL Detect Kit (ab133406) for 2 min. Band of expected size (60kDa) visualized representing strongest signal in the lane.
- Submitted by
- Noam Shoresh
- Lab
- Bradley Bernstein, Broad
- Grant
- U54HG006991
HDAC2 (Homo sapiens)
Method: ChIP-string comparison
not reviewed
- Submitted by
- Bradley Bernstein
- Lab
- Bradley Bernstein, Broad
- Grant
- U54HG004570
- Download
- human_HDAC2_validation_Bernstein.pdf
HDAC2 (Homo sapiens)
Method: immunoblot
not reviewed
- Submitted by
- Bradley Bernstein
- Lab
- Bradley Bernstein, Broad
- Grant
- U54HG004570
- Download
- human_HDAC2_validation_Bernstein.pdf