ENCGM448SCH
Summary
- Status
- released
- Description
- ChIP-Seq on HepG2
- Type
- insertion
- Tags
- 3xFLAG — C-terminal
- Purpose
- tagging
Modification site
- Target
- PHF21A-human
Modification method
- Method
- CRISPR
Documents
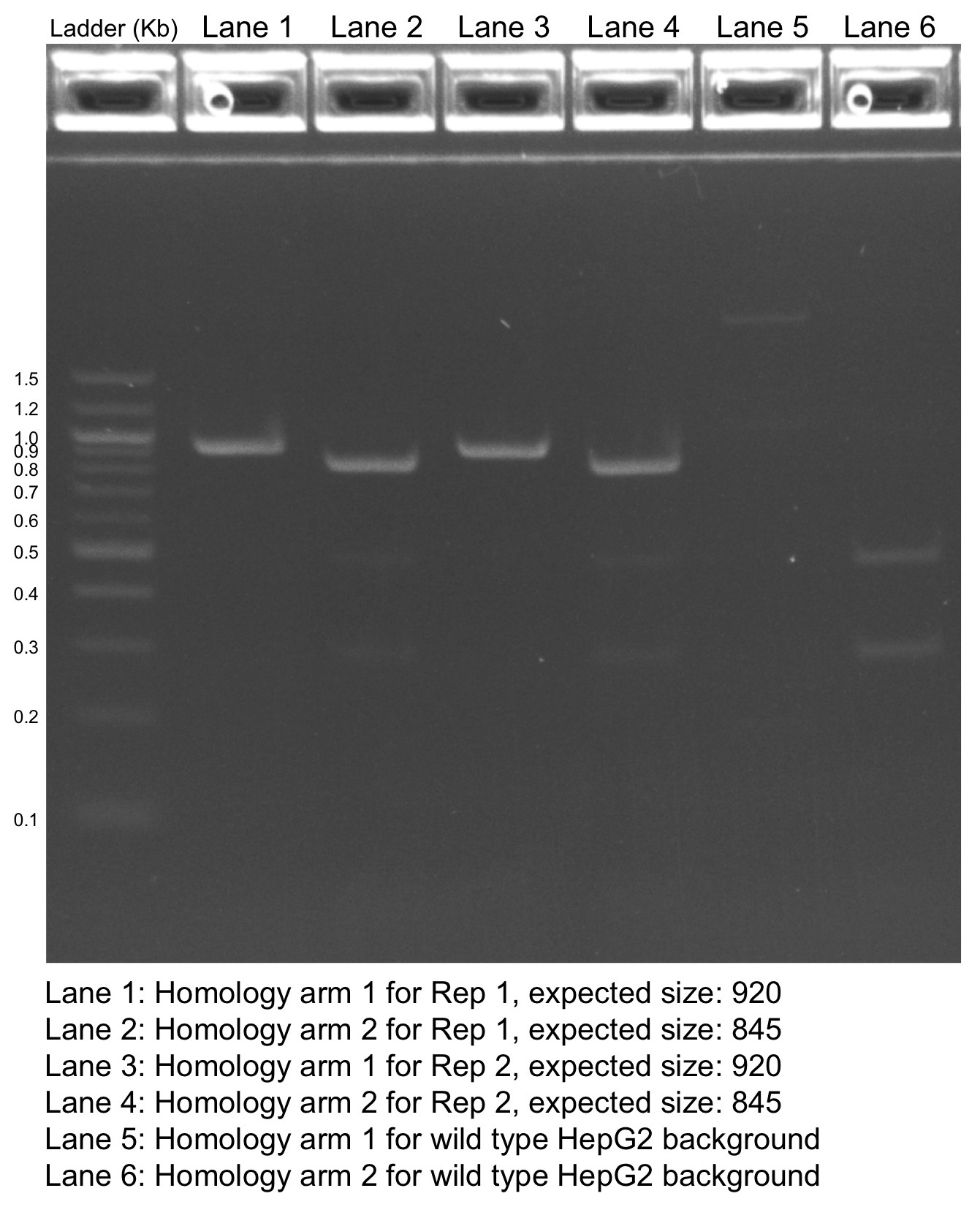
PCR analysis Characterization
- Lab
- Richard Myers, HAIB