MAD Quality Metric
Replicate log-ratio standard deviation
The Median Absolute Deviation (MAD) quality metric is more accurately described as the replicate log-ratio standard deviation. This metric evaluates reproducibility between two biological or technical replicates within RNA-seq/RAMPAGE/Entex experiments in terms of similarity of the expression levels across all pairs of genes with the present cut-off. We measure the standard deviation of the log-ratios between replicates using a robust approach, and the values are calculated per pair of replicates.
The MAD score between replicates X1 and X2 of the sample X is calculated as:
MAD = 1.4826 * median | log2(X1_i/X2_i) |
where
X1 : [X1_1,X1_2,..,X1_N] expression values in the replicate #1
X2 : [X2_1,X2_2,..,X2_N] expression values in the replicate #2
The MAD score is calculated on normalized FPKM values for genes G:1..N that have X1_i, X2_i more than preset cut-off >1 FPKM.
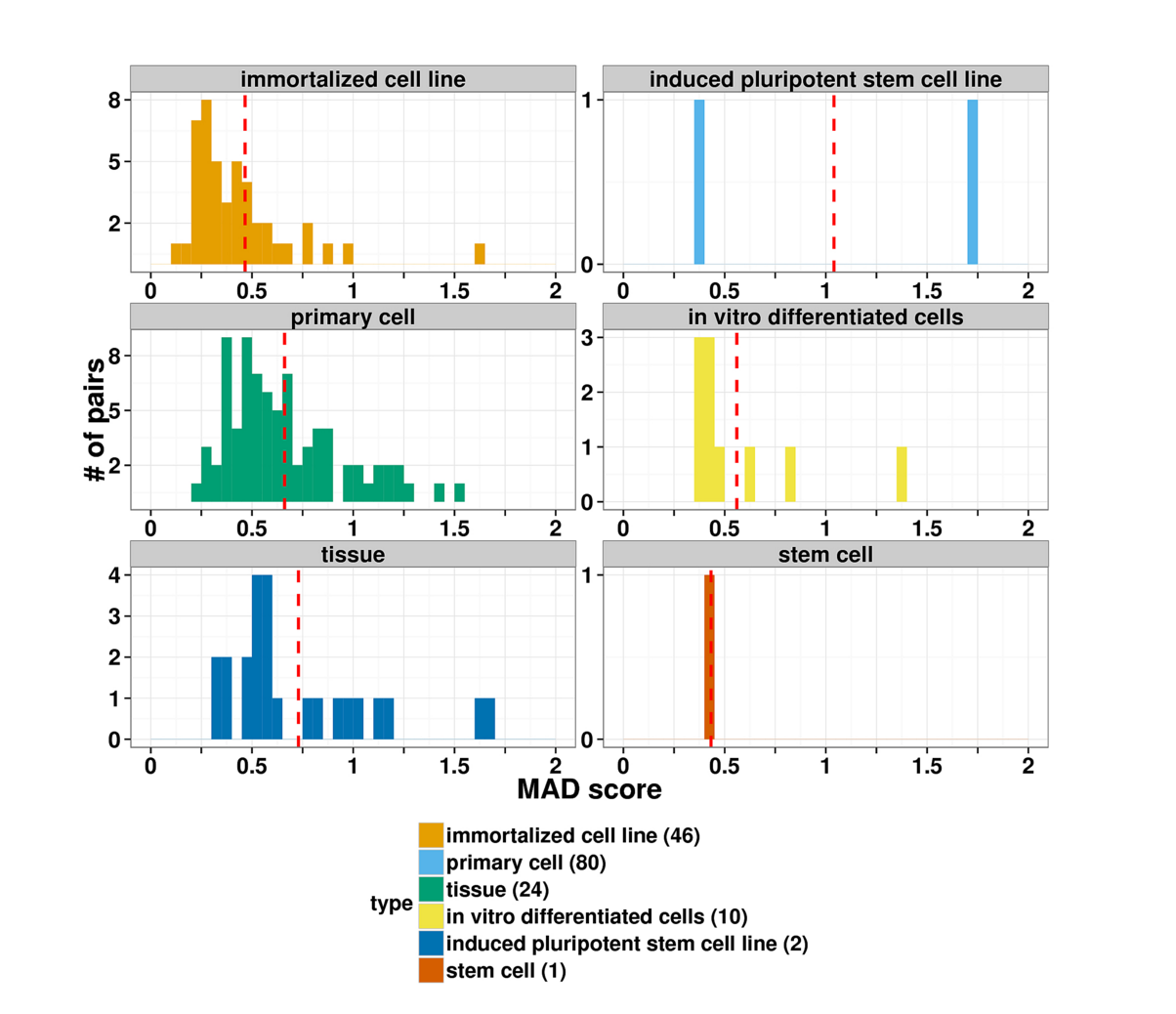
Figure 1. Distribution of the replicate log-ratio standard deviation by the sample type. The dashed red line shows mean values per each sample type; the number of the samples is indicated in the parenthesis of the legend.
References
A benchmark for RNA-seq quantification pipelines (in press)