Data Analysis
Project overview | Using the portal | REST API | Data organization | Data submission | Data analysis | Encyclopedia
ENCODE provides uniformly processed data for several major data types on the portal, including TF and Histone ChIP-seq, DNase-seq, ATAC-seq, RNA-seq, WGBS, Hi-C, and others. Detailed information on each of the available uniform processing pipelines can be accessed on the Assays and Standards page.
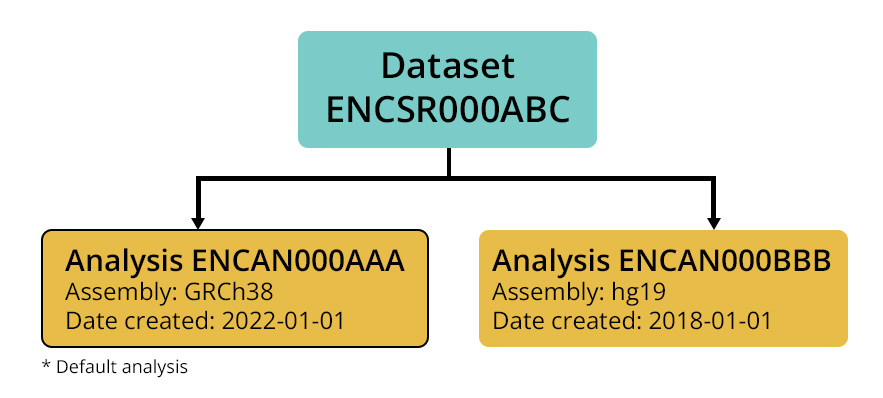
The processed data files are shared via the ENCODE Portal. Each unique processing run of a given analysis pipeline is represented by an Analysis object, which groups together all output files from that run. Each dataset is linked to all Analyses that analyzed the raw data associated with that dataset, meaning that a dataset may have multiple Analyses if it was processed multiple times (for example using different reference genome assemblies). If the data has not been processed by one of the uniform processing pipelines implemented by the DCC, the dataset may include Analyses representing processed data provided by a lab. However, datasets will always have a single “default” Analysis designated, which generally corresponds to the most recent processing run utilizing the latest version of the processing pipeline.
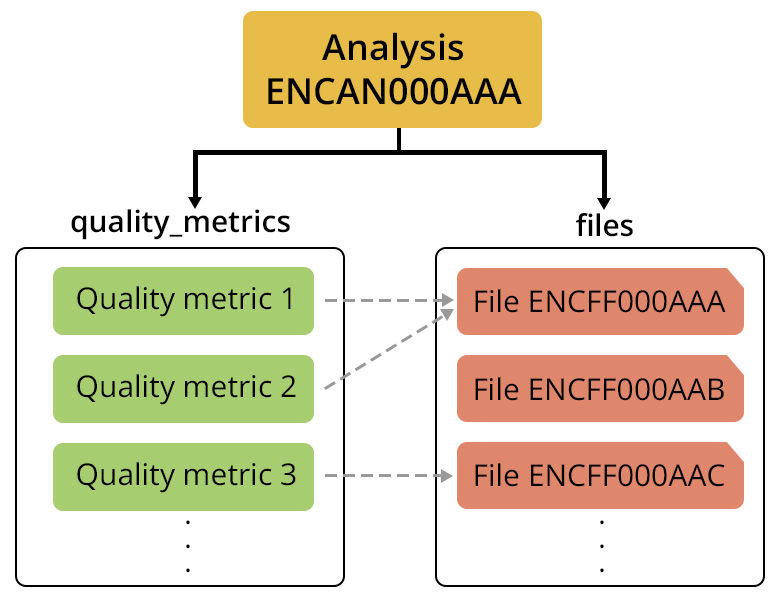
In addition to listing the output files from a pipeline run, Analyses also include relevant quality metric data in the quality_metrics property. The quality_metrics property is a list of all Quality Metric objects associated with any of the Files included in the Analysis. Each Quality Metric object contains metrics collected from the processing run, for example the number of mapped reads in an alignments file. The specific types of Quality Metric objects will depend on the data type and the pipeline used to produce the analysis. For example, the quality_metrics property of ENCAN316CWM, an Analysis of the Histone ChIP-seq dataset ENCSR707VZS, includes a ChIP Alignment Quality Metric, which is specific to ChIP-seq data.
To indicate violations of the expected quality standards thresholds, the DCC utilized an auditing system to flag relevant datasets. These audit flags appear on the dataset with a short description of the issue at hand. Detailed information on audits in general can be found at the Audits page.