ENCAB986YAH
Antibody against Homo sapiens SMARCA2
Homo sapiens
K562
characterized to standards with exemption
- Status
- released
- Source (vendor)
- Bethyl Labs
- Product ID
- A301-014A
- Lot ID
- 1
- Characterized targets
- SMARCA2 (Homo sapiens)
- Host
- rabbit
- Clonality
- polyclonal
- Purification
- affinity
- Aliases
- michael-snyder:AS-1429
- External resources
Characterizations
SMARCA2 (Homo sapiens)
K562
compliant
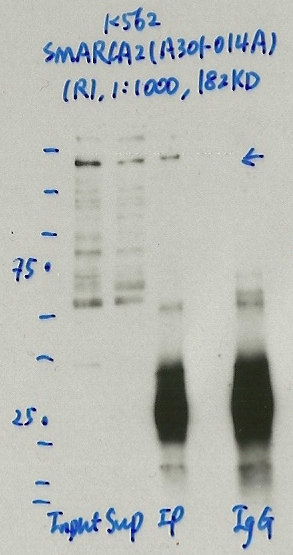
- Caption
- Immunoprecipitation was performed on nuclear extracts from the cell line: K562, using the antibody A301-014A. The blot shows western blot analysis of input, flowthrough, immunoprecipitate and mock immunoprecipitate using IgG.Molecular Weight: 181.279
- Submitted by
- Denis Salins
- Lab
- Michael Snyder, Stanford
- Grant
- U54HG006996
- Download
- 1077_01_SMARCA2_A301-014A.jpg
SMARCA2 (Homo sapiens)
K562
exempt from standards
- Caption
- Immunoprecipitation was performed on nuclear extracts from the cell line K562 using the antibody A301-014A. Lane 1: input nuclear lysate. Lane 2: material immunoprecipitated with antibody. Lane 3: material immunoprecipitated using control IgG. Marked bands were excised from gel and subjected to analysis by mass spectrometry. Target molecular weight: 181.279.
- Submitter comment
- The region corresponding to the expected size was excised for mass-spec analysis.
- Reviewer comment
- No immunoreactive signal can be seen in the submitted image but target TF is detected in downstream mass spectrometry analysis.
- Submitted by
- Nathaniel Watson
- Lab
- Michael Snyder, Stanford
- Grant
- U54HG006996
- Download
- K562-SMARCA2 (A301-014A).JPG
SMARCA2 (Homo sapiens)
Method: immunoprecipitation followed by mass spectrometry
exempt from standards
- Caption
- IP followed by mass spectrometry. Briefly, protein was immunoprecipitated from K562 nuclear cell lysates using the antibody A301-014A, and the IP fraction was loaded on a 10% polyacrylamide gel (NuPAGEBis-Tris Gel) and separated with an Invitrogen NuPAGE electrophoresis system. The gel was stained by ColloidialCoomassie G-250 stain, gel fragments corresponding to the bands indicated were excised. Then proteins were trypsinized using the in-gel digestion method. Digested proteins were analyzed on an Orbitrap Elite mass spectrometer (Thermo Scientific) by the nanoLC-ESI-MS/MS technique. Peptides were identified by the SEQUEST algorithm and filtered with a high confidence threshold (Peptide false discovery rate < 1%, 2 unique peptides per protein minimum, mass error < 10 ppm).
- Submitter comment
- MIA3 is located in endoplasmic reticulum membrane, in charge of protein transportation. All of SMARCA2, ARID1A, SMARCC1, SMARCC2, ARID1B are components of the BAF complex, http://www.genecards.org/cgi-bin/carddisp.pl?gene=SMARCA2&keywords=smarca2. Both SMARCA2 and HNRNPUL1 has interaction with EED (50KD) and EP300 (264KD), but EED and EP300 are not in the same gel slice as SMARCA2 and HNRNPUL1. MDC1 is protein C-terminus binding and FHA domain binding protein. Both SMARCA2 and YEATS2 (150KD) has interaction with TBP, http://thebiogrid.org/112479/summary/homo-sapiens/smarca2.html and http://thebiogrid.org/120815/summary/homo-sapiens/yeats2.html. TBP (38KD) is not in the same gel slice with SMARCA2 and YEATS2 . Both PBRM1 and SMARCC2 are component of the SWI/SNF-B (PBAF) chromatin-remodeling complex. http://www.genecards.org/cgi-bin/carddisp.pl?gene=PBRM1&keywords=pbrm1, http://thebiogrid.org/120490/summary/homo-sapiens/pbrm1.html. And SMARCC2 was detected and are in the BAF complex with SMARCA2.
- Reviewer comment
- SMARCA2 is not the top ranked TF in the mass spec analysis but its interacting partners in the BAF complex are ranked higher, which is acceptable according to the ENCODE antibody standards document.
- Submitted by
- Nathaniel Watson
- Lab
- Michael Snyder, Stanford
- Grant
- U54HG006996
- Download
- SMARCA2_A301-014A_final.pdf