ENCAB728YTO
Antibody against Aequorea victoria eGFP
Caenorhabditis elegans
whole organism
characterized to standards
Homo sapiens
K562, MCF-7, GM23338, A549, SK-N-SH, HepG2, GM12878, WTC11, glutamatergic neuron
characterized to standards
Drosophila melanogaster
whole organism
characterized to standards
Homo sapiens
excitatory neuron
awaiting characterization
- Status
- released
- Source (vendor)
- Kevin White
- Product ID
- goat-anti-GFP-UC
- Lot ID
- unknown
- Characterized targets
- eGFP (Aequorea victoria)
- Host
- goat
- Clonality
- polyclonal
- Purification
- affinity
- Antigen description
- Antisera raised against a peptide from the GFP protein. Produced commercially for the modENCODE Project by K. White's group and Millipore.
- Antigen sequence
- MSKGEELFTGVVPILVELDGDVNGHKFSVSGEGEGDATYGKLTLKFICTTGKLPVPWPTLVTTLTYGVQCFSRYPDHMKRHDFFKSAMPEGYVQERTIFFKDDGNYKTRAEVKFEGDTLVNRIELKGIDFKEDGNILGHKLEYNYNSHNVYIMADKQKNGIKVNFKIRHNIEDGSVQLADHYQQNTPIGDGPVLLPDNHYLSTQSALSKDPNEKRDHMVLLEFVTAAGITHGMDELYK
- Aliases
- kevin-white:GFP NewGoat UofC, kevin-white:GoatV-anti-GFP
- External resources
Characterizations
eGFP (Aequorea victoria)
A549
compliant
- Caption
- Immunoprecipitation was performed on nuclear extracts of eGFP-FOSL2 tagged A549 cells using anti-GFP antibody. The image shows western blot analysis of input, flow-through, immunoprecipitate, and mock immunoprecipitate using IgG. Molecular weight standard contains 10 pre-stained recombinant proteins (250, 150, 100, 75, 50, 37, 25, 20, 15, and 10 kD). Target molecular weight is 63 kD.
- Reviewer comment
- UniProt (P15408) lists 3 known isoforms of FOSL2 in the 30-35kDa range with the addition of GFP (~27 kDa), is consistent with the size seen in the blot.
- Submitted by
- Jessika Adrian
- Lab
- Michael Snyder, Stanford
- Grant
- UM1HG009442
- Download
- 11_A549_FOSL2-500-Rep1_IP.jpg
eGFP (Aequorea victoria)
MCF-7
compliant
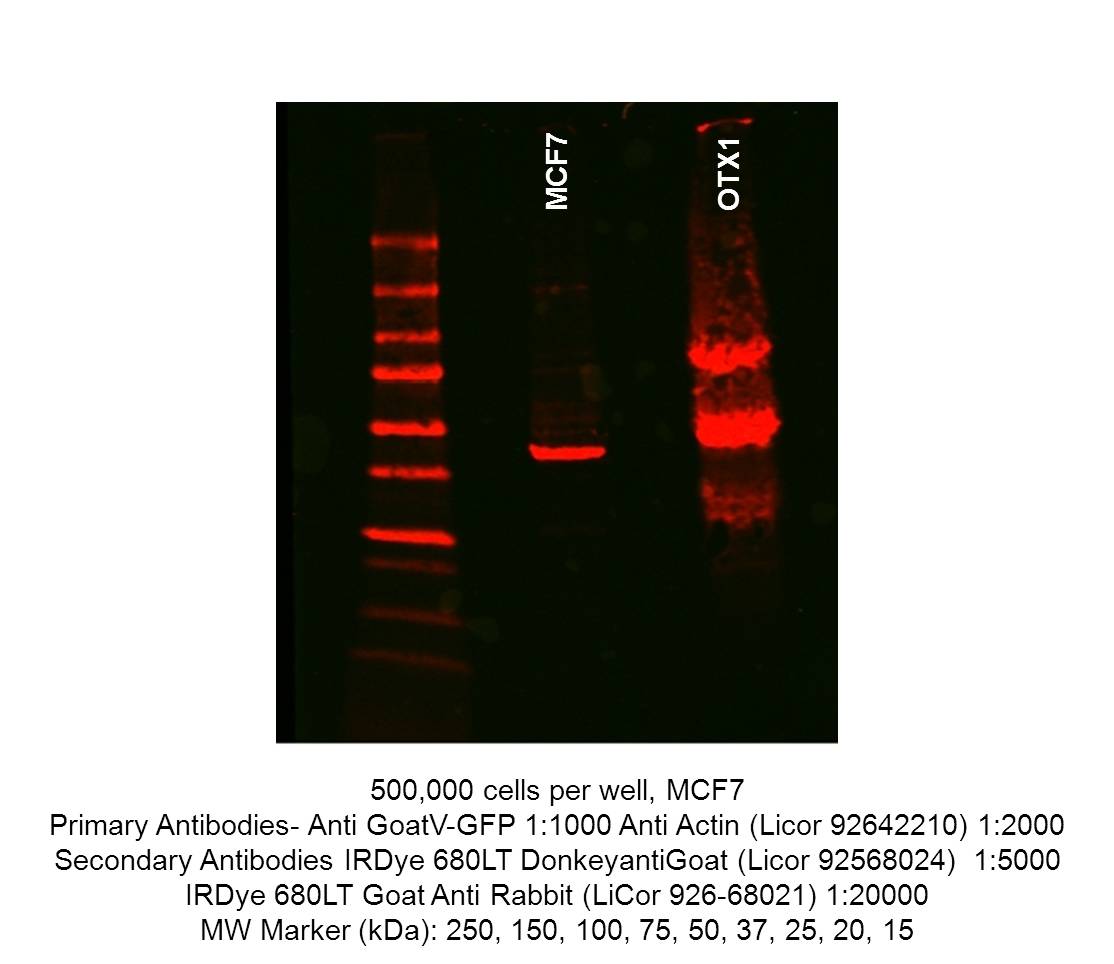
- Caption
- Immunoblot of untransfected MCF7 and OTX-1-GFP tagged MCF7. 500,000 cells were resuspended in 250μL cold RIPA buffer(EMD Millipore 20-188), incubated on ice for 15 mineuts prior to centrifugation at 4°C, 14,000g for 10 minutes. In a new tube, 250μL Laemmli sample buffer(Bio-rad 161-0737) and 50μL β-mercaptoethanol was added to the supernatant. The samples were incubated at 100°C for 10 minutes, before cooling and storage at -20°C. 40uL of lysate was run on precast Mini-PROTEAN TGX gels (Biorad 4561094) and transfered to nitrocellulose membranes. The western blot blocked in Odessey Blocking buffer (Licor 92740000) and incubated with GoatV-GFP (1:1000) and anti-actin(Licor 92642210, 1:2000). The secondary antibody is donkey anti-goat (Licor 92568024, 1:5000).
- Reviewer comment
- The signal in the untransfected MCF-7 lane is supposedly from the secondary antibody cross-reacting with actin according to the submitting lab.
- Submitted by
- Alec Victorsen
- Lab
- Kevin White, UChicago
- Grant
- U41HG007355
- Download
- 20160330_MCF7_GoatV_control.jpg
eGFP (Aequorea victoria)
GM23338
compliant
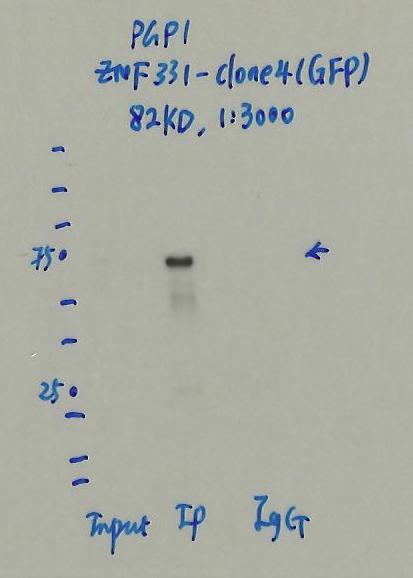
- Caption
- Immunoprecipitation was performed on nuclear extracts of eGFP-ZNF331 tagged PGP1 cells using anti-GFP antibody. The image shows western blot analysis of input, flow-through, immunoprecipitate, and mock immunoprecipitate using IgG. Molecular weight standard contains 10 pre-stained recombinant proteins (250, 150, 100, 75, 50, 37, 25, 20, 15, and 10 kD). Target molecular weight is 82 kD.
- Submitter comment
- Detected band is within 20% of expected target size.
- Submitted by
- Jessika Adrian
- Lab
- Michael Snyder, Stanford
- Grant
- UM1HG009442
- Download
- 22_PGP1_ZNF331_clone4.jpg
eGFP (Aequorea victoria)
SK-N-SH
compliant
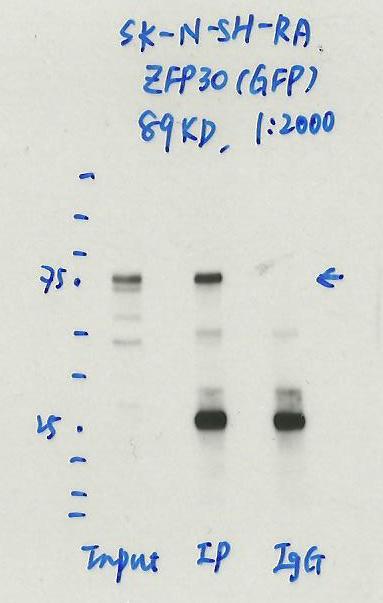
- Caption
- Immunoprecipitation was performed on nuclear extracts of RA treated eGFP-ZFP30 tagged SK-N-SH cells using anti-GFP antibody. The image shows western blot analysis of input, flow-through, immunoprecipitate, and mock immunoprecipitate using IgG. Molecular weight standard contains 10 pre-stained recombinant proteins (250, 150, 100, 75, 50, 37, 25, 20, 15, and 10 kD). Target molecular weight is 89 kDa.
- Reviewer comment
- ZFP30 is ~61.5kDa according to UniProt (Q9Y2G7) and eGFP is ~27kDa, which places the immunoreactive band around the expected size.
- Submitted by
- Jessika Adrian
- Lab
- Michael Snyder, Stanford
- Grant
- UM1HG009442
- Download
- 8_SK-N-SH-RA_ZNF30_IP.jpg
eGFP (Aequorea victoria)
compliant
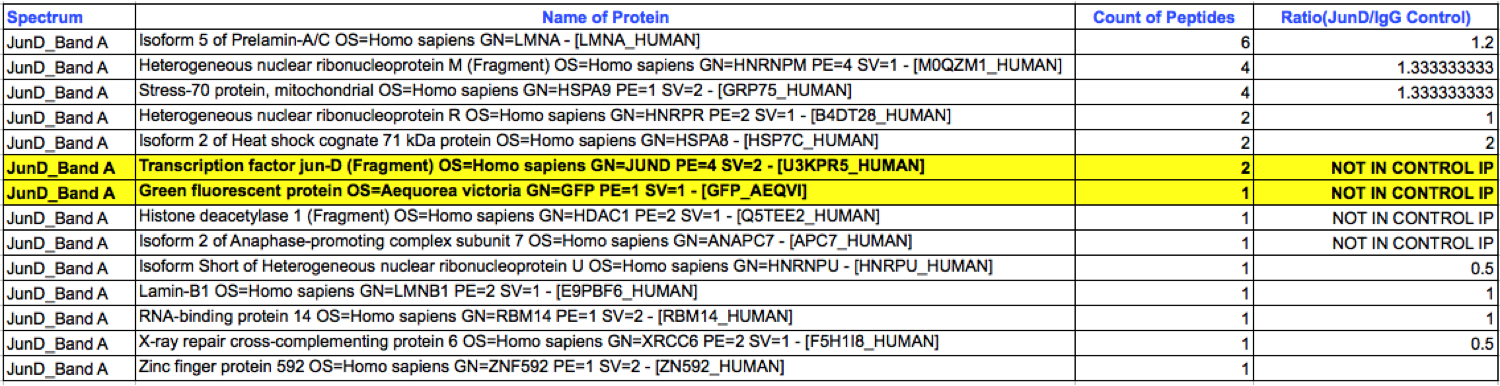
- Caption
- K562 whole cell lysates were immunoprecipitated using anti-GFP antibody and the IP fraction was loaded on a 10% polyacrylamide gel (NuPAGE Bis-Tris Gel) and separated with an invitrogen NuPAGE electrophoresis system. The gel was silver-stained, gel fragments corresponding to the bands indicated were excised and destained using the SilverSNAP Stain for Mass Spectrometry (Pierce). Then proteins were trysinized using the in-gel digestion method. Digested proteins were analyzed on an LTQ-Orbitrap (Thermo Scientific) by the nanoLC-ESI-MS/MS technique. Peptides were identified by the SEQUEST algorithm and filtered with a high confidence threshold (Protein false discovery rate < 1%, 2 peptides per protein minimum). The target protein, JunD, was identified as the 7th ranked enriched protein based on IP-Mass Spectrometry.
- Submitted by
- Minyi Shi
- Lab
- Michael Snyder, Stanford
- Grant
- U54HG006996
- Download
- eGFP_ENCAB728YTO_IP-MS.png
eGFP (Aequorea victoria)
whole organism
compliant
- Caption
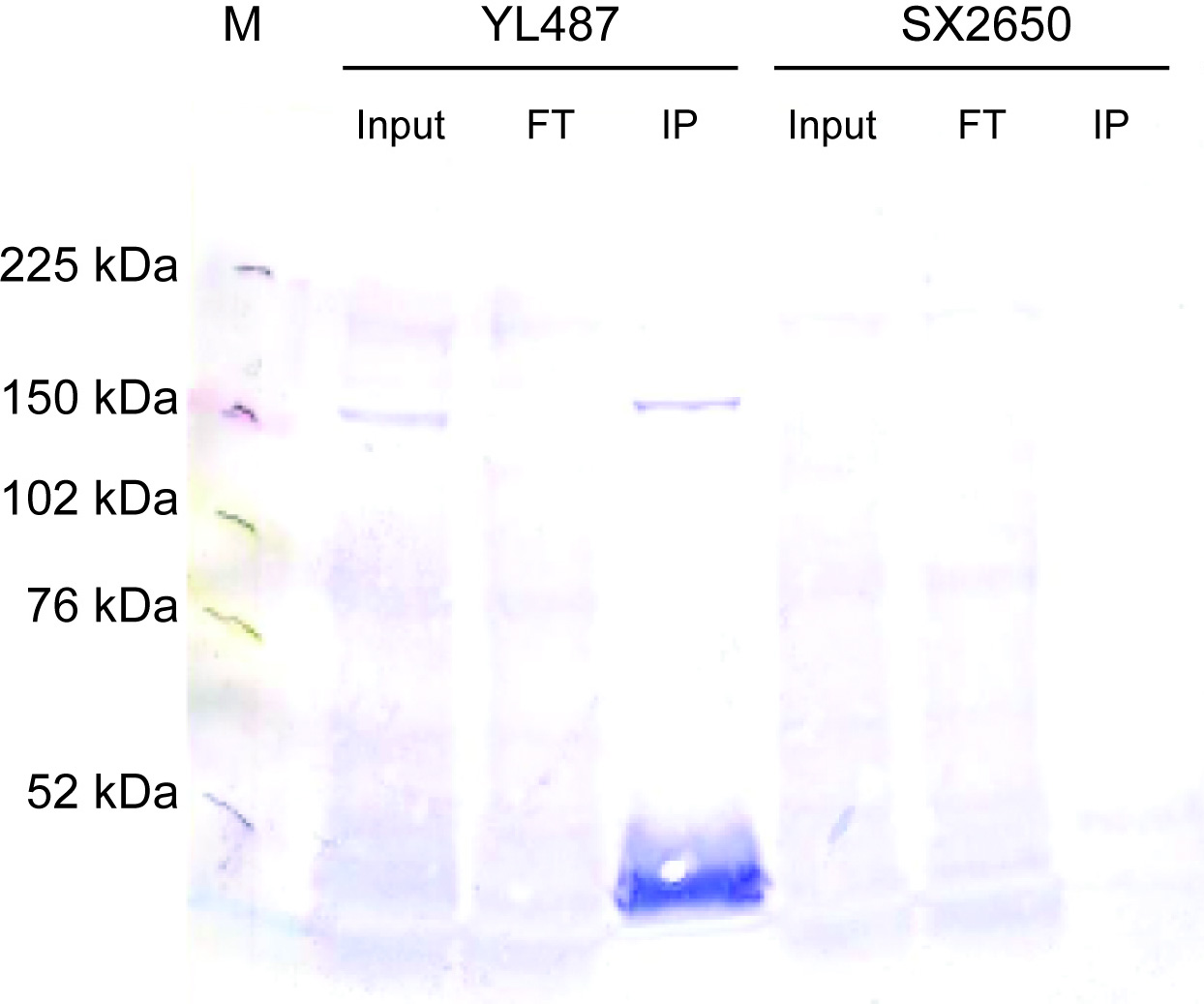
- ChIP assays were conducted as previously described (Zhong et al., 2010). Young adults were collected from YL487 (snpc-4(tm4568) I; unc-119(ed3)III; wgIs179 [snpc-4:TY1 EGFP 3XFLAG; unc-119]). 2.2 mg of cell extract was immunoprecipitated using 7.5 μg of anti-GFP antibody. Ten percent of input and flow-through as well as the IP were loaded onto a 6% SDS-PAGE gel. The gel was transferred onto PVDF, rehydrated in methanol and Ponceau stained. After blocking, the blot was incubated with anti-FLAG M2 (Sigma F1804) at 1:1000 in 2.5% milk/TBST overnight at 4 degrees. After washing in TBST, the blot was incubated with anti-mouse IgG-AP (1:5000) in TBST for 1 hour at room temperature and then washed before developing with alkaline phosphatase.
- Reviewer comment
- As stated in the August 2015 antibody characterization standards document, the requirements for epitope-tagged proteins are described elsewhere (i.e. in the July 2011 ChIP-seq standards document). On page 4, it states "The recommended control for experiments with epitope-tagged factors is an immunoprecipitation using the same antibody against the epitope tag in otherwise identical cells which do not contain the expression construct or which do not express the tagged factor." Hence, an IgG-control is not needed.
- Submitted by
- Alec Victorsen
- Lab
- Kevin White, UChicago
- Grant
- U41HG007355
- Download
- GoatV verification blot.jpg
eGFP (Aequorea victoria)
whole organism
compliant
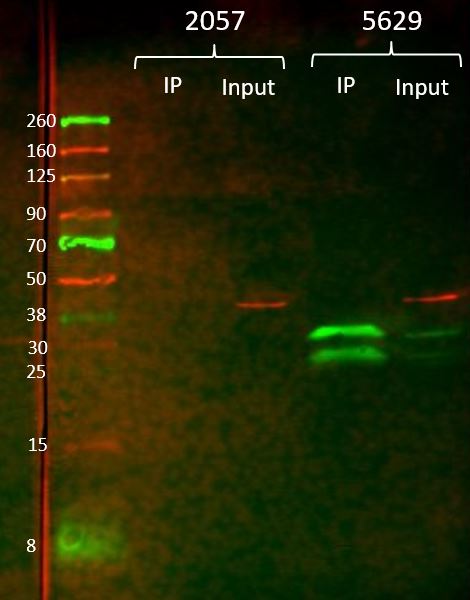
- Caption
- For the immunoprecipitation, ~15 ug anti-GFP (GFPV) antibody was crosslinked to 100 uL gammabind plus sepharose beads (GE). The antibody-bead conjugate was then incubated overnight at 4 degC with 100 mg of whole animal extract from flies overexpressing GFP (BDSC #5629) or non-transgenic flies (BDSC #2057) in 750uL T-PER (Fisher). The samples were washed 3x with lysis buffer (DCC modENCODE protocol) and eluted from the beads with 100 uL 1M glycine pH 3. The samples were neutralized with 100 uL Tris-HCL pH 8. Input samples (5uL) and IP samples (20uL) were diluted with 2x Laemmli buffer and run on a mini-PROTEAN gel (bio-rad) and transferred to a nitrocellulose membrane. The membrane was incubated with Roche anti-GFP (1:1000) and anti-actin (1:2000) primary antibodies overnight at 4 degC, then Licor IR680 (1:20000) and IR800 (1:5000) secondary antibodies for 1 hour at RT. After washing, the membrane was imaged on a Licor Odyssey scanner. The 34.8kDa band represents the GFP with a nuclear localization signal. The 27kDa band is free GFP.
- Reviewer comment
- As stated in the August 2015 antibody characterization standards document, the requirements for epitope-tagged proteins are described elsewhere (i.e. in the July 2011 ChIP-seq standards document). On page 4, it states "The recommended control for experiments with epitope-tagged factors is an immunoprecipitation using the same antibody against the epitope tag in otherwise identical cells which do not contain the expression construct or which do not express the tagged factor." Hence, an IgG-control is not needed.
- Submitted by
- Alec Victorsen
- Lab
- Kevin White, UChicago
- Grant
- U41HG007355
- Download
- goatV-fly-pri_char.jpg
eGFP (Aequorea victoria)
GM12878
not compliant
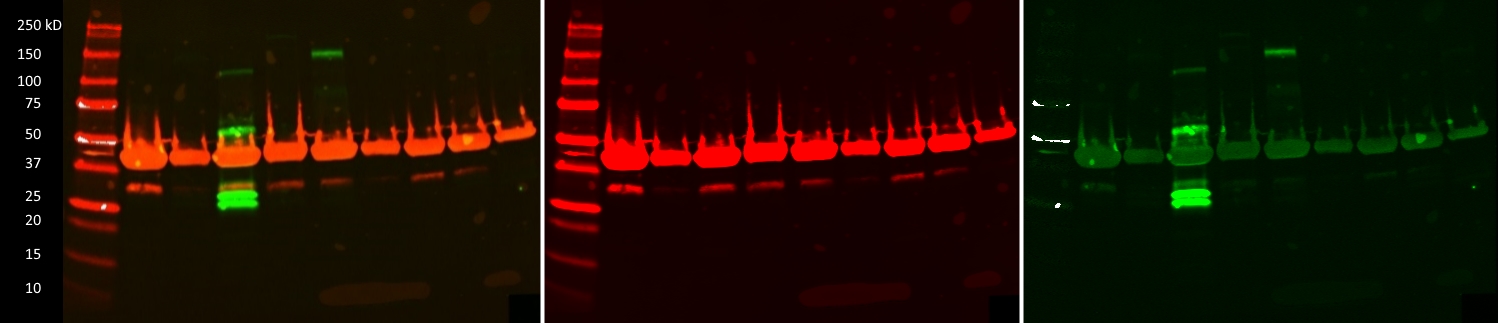
- Caption
- IP-Western images of 9 GM12878 lines transfected with CRISPR constructs designed to create GFP tagged Transcription Factors. The targeted TFs from left to right are ATF4, NR2F6, PRDM15, ZBTB4, ZBTB17, ZNF215, ZNF561, ZNF641 and ZSCAN32. Only PRDM15 (~204, 170, 95kD) and ZBTB17 (~122, 114kD) appear to be GFP positive. Combined blot image shows actin signal in red and GFP signal in green. Extracts from potentially transgenic cells were IPed using the goatV anti-GFP antibody (ENCAB728YTO). The western blot was incubated with anti-GFP (mouse, 1:5000, Roche p/n: 11814460001) and anti-actin (rabbit, 1:2000, Licor p/n: 926-42210) antibodies. After secondary incubation with Licor IR800CW (goat anti-mouse, 1:5000) and Licor IR680LT (goat anti-rabbit, 1:20000) the blot was imaged at 700 and 800nm (2nd and 3rd images respectivly).
- Submitter comment
- Unlike the ZBTB17-GFP sample, the most intense bands for PRDM15-GFP are well below the predicted size of the fusion protein. However, given they are absent in the other TF-GFP lines, they are specific to the PRDM15-GFP. Given this specificity, as well as the fact that these low molecular weight bands are roughly the same size as the GFP epitope alone, we believe that these bands still represent GFP signal. Even after further validation, which may prove PRDM15-GFP to be a valid ENCODE CRISPR tagged line, these additional bands could occur for any number of legitimate post-translational or experimental means
- Reviewer comment
- The major band in the PRDM15 lane is much lower than the expected size of the known protein (UniProt: http://www.uniprot.org/uniprot/P57071) isoforms (~57kDa to ~169kDa) and is much more consistent with the size of the eGFP tag only. For the ZBTB17 lane, the major band is too high relative to the expected size of the known isoforms (UniProt:http://www.uniprot.org/uniprot/Q13105, 78-88kDa range), not within the allowed 20% size deviation according to the ENCODE standards.
- Submitted by
- Alec Victorsen
- Lab
- Kevin White, UChicago
- Grant
- UM1HG009442
- Download
- GoatV_GM12878.jpg
eGFP (Aequorea victoria)
HepG2
compliant
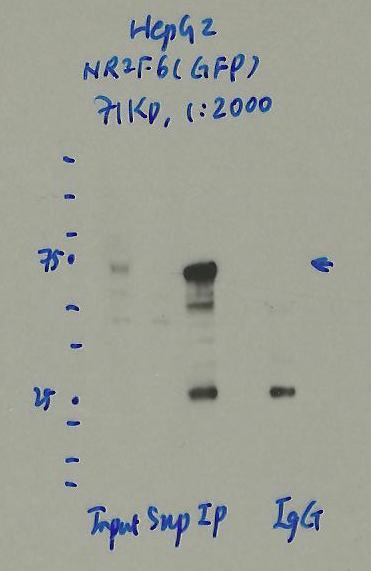
- Caption
- Immunoprecipitation was performed on nuclear extracts of eGFP-NR2F6 tagged HepG2 cells using anti-GFP antibody. The image shows western blot analysis of input, flow-through, immunoprecipitate, and mock immunoprecipitate using IgG. Molecular weight standard contains 10 pre-stained recombinant proteins (250, 150, 100, 75, 50, 37, 25, 20, 15, and 10 kD). Target molecular weight is 71 kDa.
- Reviewer comment
- NR2F6 is ~43kDa according to UniProt (P10588) and eGFP is ~27kDa which places the immunoreactive band around the expected size.
- Submitted by
- Jessika Adrian
- Lab
- Michael Snyder, Stanford
- Grant
- UM1HG009442
- Download
- HepG2_NR2F6_IP.jpg
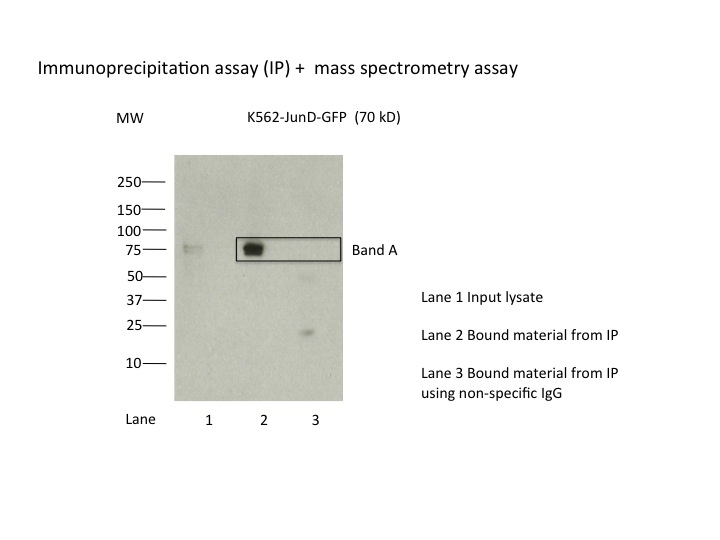
eGFP (Aequorea victoria)
K562
compliant
- Caption
- 8.3 X10^6 of cell nuclear extract was immunoprecipitated using 2µg of anti-GFP antibody and 2µg of goat IgG, respectively. Seven percent of input and flow-through as well as the IP were loaded onto a 4-15% SDS-PAGE gel. The gel was transferred onto PVDF membrane. After blocking for 1 hour at 4C, the blot was incubated with anti-GFP antibody at 1:1000 in 5% milk/PBST overnight at 4C. After washing in PBST, the blot was incubated with light chain specific anti-goat IgG-HRP (1:10000) in 5%milk/PBST for 2 hours at 4C. After washing 3 times with PBST, the blot was developed with chemiluminescent substrate.
- Submitted by
- Trupti Kawli
- Lab
- Michael Snyder, Stanford
- Grant
- U54HG006996
- Download
- K562_JunD_GFP.jpg
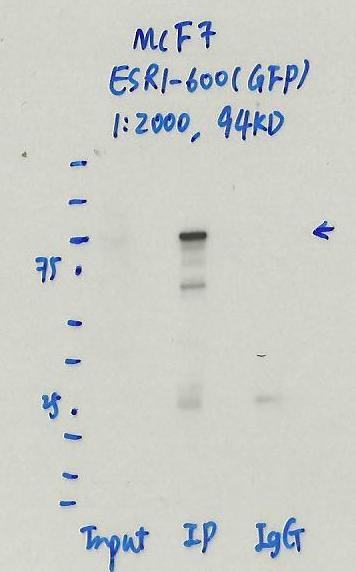
eGFP (Aequorea victoria)
MCF-7
not submitted for review by lab
- Caption
- Immunoprecipitation was performed on nuclear extracts of eGFP-ESR1 tagged MCF-7 cells using anti-GFP antibody. The image shows western blot analysis of input, flow-through, immunoprecipitate, and mock immunoprecipitate using IgG. Molecular weight standard contains 10 pre-stained recombinant proteins (250, 150, 100, 75, 50, 37, 25, 20, 15, and 10 kD). Target molecular weight is 94 kDa.
- Submitted by
- Jessika Adrian
- Lab
- Michael Snyder, Stanford
- Grant
- UM1HG009442
- Download
- MCF7_ESR1-600_IP.jpg