ENCAB320XLX
Antibody against Homo sapiens TBX3
Homo sapiens
HepG2
characterized to standards with exemption
- Status
- released
- Source (vendor)
- Bethyl Labs
- Product ID
- A303-098A
- Lot ID
- 1
- Characterized targets
- TBX3 (Homo sapiens)
- Host
- rabbit
- Clonality
- polyclonal
- Purification
- affinity
- Aliases
- michael-snyder:AS-1033
- External resources
Characterizations
TBX3 (Homo sapiens)
HepG2
not compliant
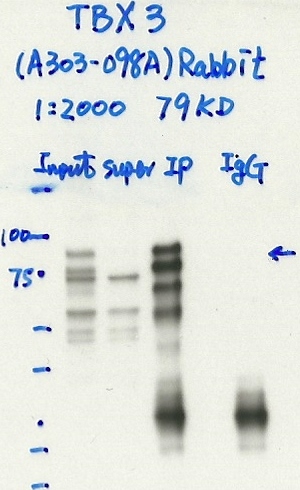
- Caption
- Immunoprecipitation was performed on nuclear extracts from the cell line: HepG2, using the antibody A303-098A. The blot shows western blot analysis of input, flowthrough, immunoprecipitate and mock immunoprecipitate using IgG.
- Reviewer comment
- Multiple bands detected and indicated size not at least 50% of the signal. Could potentially be rescued with a knockdown characterization.
- Submitted by
- Denis Salins
- Lab
- Michael Snyder, Stanford
- Grant
- U54HG006996
- Download
- Expt855_3-TBX3.jpg
TBX3 (Homo sapiens)
HepG2
exempt from standards
- Caption
- Immunoprecipitation was performed on nuclear extracts from the cell line HepG2 using the antibody A303-098A. Lane 1: input nuclear lysate. Lane 2: material immunoprecipitated with antibody. Lane 3: material immunoprecipitated using control IgG. Marked bands were excised from gel and subjected to analysis by mass spectrometry. Target molecular weight: 79.389.
- Submitter comment
- We analyzed multiple immunoreactive bands by mass-spec.
- Reviewer comment
- Multiple bands but mass spec analysis showed the target TF to be highest ranked and not in IgG with no other site specific TFs above the target.
- Submitted by
- Nathaniel Watson
- Lab
- Michael Snyder, Stanford
- Grant
- U54HG006996
- Download
- MS1036_5_TBX3-A303-098A.JPG
TBX3 (Homo sapiens)
Method: immunoprecipitation followed by mass spectrometry
exempt from standards
- Caption
- IP followed by mass spectrometry. Briefly, protein was immunoprecipitated from HepG2 nuclear cell lysates using the antibody A303-098A, and the IP fraction was loaded on a 10% polyacrylamide gel (NuPAGEBis-Tris Gel) and separated with an Invitrogen NuPAGE electrophoresis system. The gel was stained by ColloidialCoomassie G-250 stain, gel fragments corresponding to the bands indicated were excised. Then proteins were trypsinized using the in-gel digestion method. Digested proteins were analyzed on an Orbitrap Elite mass spectrometer (Thermo Scientific) by the nanoLC-ESI-MS/MS technique. Peptides were identified by the SEQUEST algorithm and filtered with a high confidence threshold (Peptide false discovery rate < 1%, 2 unique peptides per protein minimum, mass error < 10 ppm).
- Submitter comment
- We can not explain the higher ranked TFs.
- Reviewer comment
- PF: I’m not worried about YLPM1 and TAF15 because there is no evidence that these are likely to be site-specific TFs. But the issue is ZNF687- this one has more peptides than the target TF but neither are detected in the control IgG. We can’t say that it is not compliant since there are no peptides in the control IgG.
- Submitted by
- Nathaniel Watson
- Lab
- Michael Snyder, Stanford
- Grant
- U54HG006996
- Download
- TBX3_A303-098A_final.pdf