ENCAB247ONX
Antibody against Homo sapiens NFRKB
Homo sapiens
HepG2
characterized to standards
- Status
- released
- Source (vendor)
- Sigma
- Product ID
- HPA007128
- Lot ID
- R04503
- Characterized targets
- NFRKB (Homo sapiens)
- Host
- rabbit
- Clonality
- polyclonal
- Purification
- affinity
- Isotype
- IgG
- Antigen description
- Nuclear factor related to κ-B-binding protein recombinant protein epitope signature tag (PrEST)
- Antigen sequence
- KSSSGVLLVSSPTMPHLGTMLSPASSQTAPSSQAAARVVSHSGSAGLSQVRVVAQPSLPAVPQQSGGPAQTLPQMPAGPQIRVPATATQTKVVPQTVMATVPVKAQTTAATV
- Aliases
- michael-snyder:749
- External resources
Characterizations
NFRKB (Homo sapiens)
HepG2
compliant
- Caption
- Immunoprecipitation was performed on nuclear extracts from the cell line: HepG2, using the antibody HPA007128. The blot shows western blot analysis of input, flowthrough, immunoprecipitate and mock immunoprecipitate using IgG.
- Submitted by
- Denis Salins
- Lab
- Michael Snyder, Stanford
- Grant
- U54HG006996
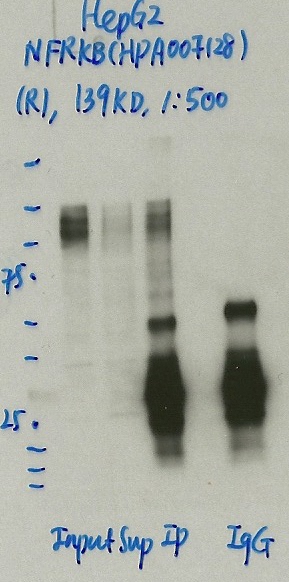
NFRKB (Homo sapiens)
HepG2
compliant
- Caption
- Immunoprecipitation was performed on nuclear extracts from the cell line HepG2 using the antibody HPA007128. Lane 1: input nuclear lysate. Lane 2: material immunoprecipitated with antibody. Lane 3: material immunoprecipitated using control IgG. Marked bands were excised from gel and subjected to analysis by mass spectrometry. Target molecular weight: 139.001.
- Submitted by
- Nathaniel Watson
- Lab
- Michael Snyder, Stanford
- Grant
- U54HG006996
- Download
- MS1036_2_NFRKB-HPA007128.JPG
NFRKB (Homo sapiens)
Method: immunoprecipitation followed by mass spectrometry
compliant
- Caption
- IP followed by mass spectrometry. Briefly, protein was immunoprecipitated from HepG2 nuclear cell lysates using the antibody HPA007128, and the IP fraction was loaded on a 10% polyacrylamide gel (NuPAGEBis-Tris Gel) and separated with an Invitrogen NuPAGE electrophoresis system. The gel was stained by ColloidialCoomassie G-250 stain, gel fragments corresponding to the bands indicated were excised. Then proteins were trypsinized using the in-gel digestion method. Digested proteins were analyzed on an Orbitrap Elite mass spectrometer (Thermo Scientific) by the nanoLC-ESI-MS/MS technique. Peptides were identified by the SEQUEST algorithm and filtered with a high confidence threshold (Peptide false discovery rate < 1%, 2 unique peptides per protein minimum, mass error < 10 ppm).
- Submitter comment
- None of the proteins ranked above or equally ranked have been shown to be sequence-specific TFs (including ILF3, EWSR1).
- Submitted by
- Nathaniel Watson
- Lab
- Michael Snyder, Stanford
- Grant
- U54HG006996
- Download
- NFRKB_HPA007128_final.pdf