ENCAB125OWA
Alternate accession: ENCAB000AEL
Antibody against Homo sapiens BRCA1
Homo sapiens
K562, HeLa-S3
characterized to standards
Homo sapiens
any cell type or tissue
partially characterized
Homo sapiens
GM12878, HepG2
not characterized to standards
- Status
- released
- Source (vendor)
- Bethyl Labs
- Product ID
- A300-000A
- Lot ID
- 2
- Characterized targets
- BRCA1 (Homo sapiens)
- Host
- rabbit
- Clonality
- polyclonal
- Aliases
- bradley-bernstein:PchAb 844
- External resources
Characterizations
BRCA1 (Homo sapiens)
not submitted for review by lab
- Caption
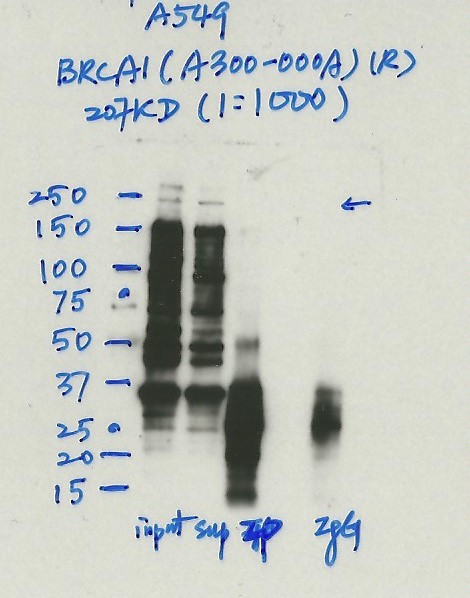
- Immunoprecipitation was performed on nuclear extracts from the cell line: A549, using the antibody A300-000A. The blot shows western blot analysis of input, flowthrough, immunoprecipitate and mock immunoprecipitate using IgG.Molecular Weight: 207.7
- Submitted by
- Nathaniel Watson
- Lab
- Michael Snyder, Stanford
- Grant
- U54HG006996
- Download
- #1132A549 BRCA1(A300-000A) (2).jpg
BRCA1 (Homo sapiens)
not submitted for review by lab
- Submitted by
- Noam Shoresh
- Lab
- Bradley Bernstein, Broad
- Grant
- U54HG006991
- Download
- BRCA1_Bethyl_A300-000A-2_WB.png
BRCA1 (Homo sapiens)
Method: immunoprecipitation followed by mass spectrometry
compliant
- Caption
- IP followed by mass spectrometry: Briefly, protein was immunoprecipitated from K562 whole cell lysates using A300-000A, and the IP fraction was loaded on a 10% polyacrylamide gel (NuPAGE Bis-Tris Gel) and separated with an Invitrogen NuPAGE electrophoresis system. The gel was silver-stained, gel fragments corresponding to the bands indicated were excised and destained using the SilverSNAP Stain for Mass Spectrometry (Pierce). Then proteins were trypsinized using the in-gel digestion method. Digested proteins were analyzed on an LTQ-Orbitrap (Thermo Scientific) by the nanoLC-ESI-MS/MS technique. Peptides were identified by the SEQUEST algorithm and filtered with a high confidence threshold (Protein false discovery rate < 1%, 2 peptides per protein minimum). Based on these observations, this band is likely due to the presence of immunoprecipitated BRCA1 and A300-000A meets the ENCODE standard for validation by this criterion.
- Submitted by
- Kathrina Onate
- Lab
- Michael Snyder, Stanford
- Grant
- U54HG004558
- Download
- BRCA1_final_AEL Sheet1.pdf
BRCA1 (Homo sapiens)
not submitted for review by lab
- Caption
- Immunoprecipitation was performed on nuclear extracts from the cell line: HEK293T, using the antibody A300-000A. The blot shows western blot analysis of input, flowthrough, immunoprecipitate and mock immunoprecipitate using IgG.Molecular Weight: 207.7
- Submitted by
- Nathaniel Watson
- Lab
- Michael Snyder, Stanford
- Grant
- U54HG006996
- Download
- Expt1118_3-BRCA1-A300-000A.JPG
BRCA1 (Homo sapiens)
HeLa-S3
Method: immunoblot
compliant
- Caption
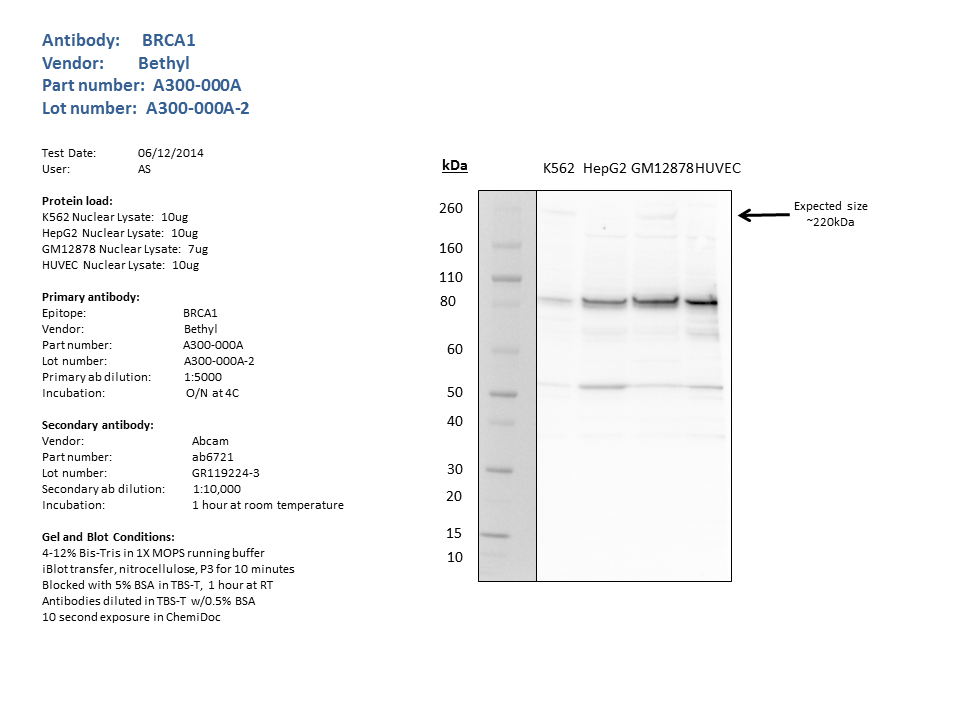
- Western blots on nuclear lysates from cell lines GM12878 (Lane1), K562 (Lane2), HeLaS3 (Lane3), and HepG2 (Lane4). A band of ~207kD is detected by Western blotting with A300-000A in K562 and HelaS3 nuclear lysates
- Submitted by
- Michael Snyder
- Lab
- Michael Snyder, Stanford
- Grant
- U54HG004558
- Download
- human_BRCA1_validation_Snyder.pdf
BRCA1 (Homo sapiens)
Method: immunoprecipitation followed by mass spectrometry
compliant
- Caption
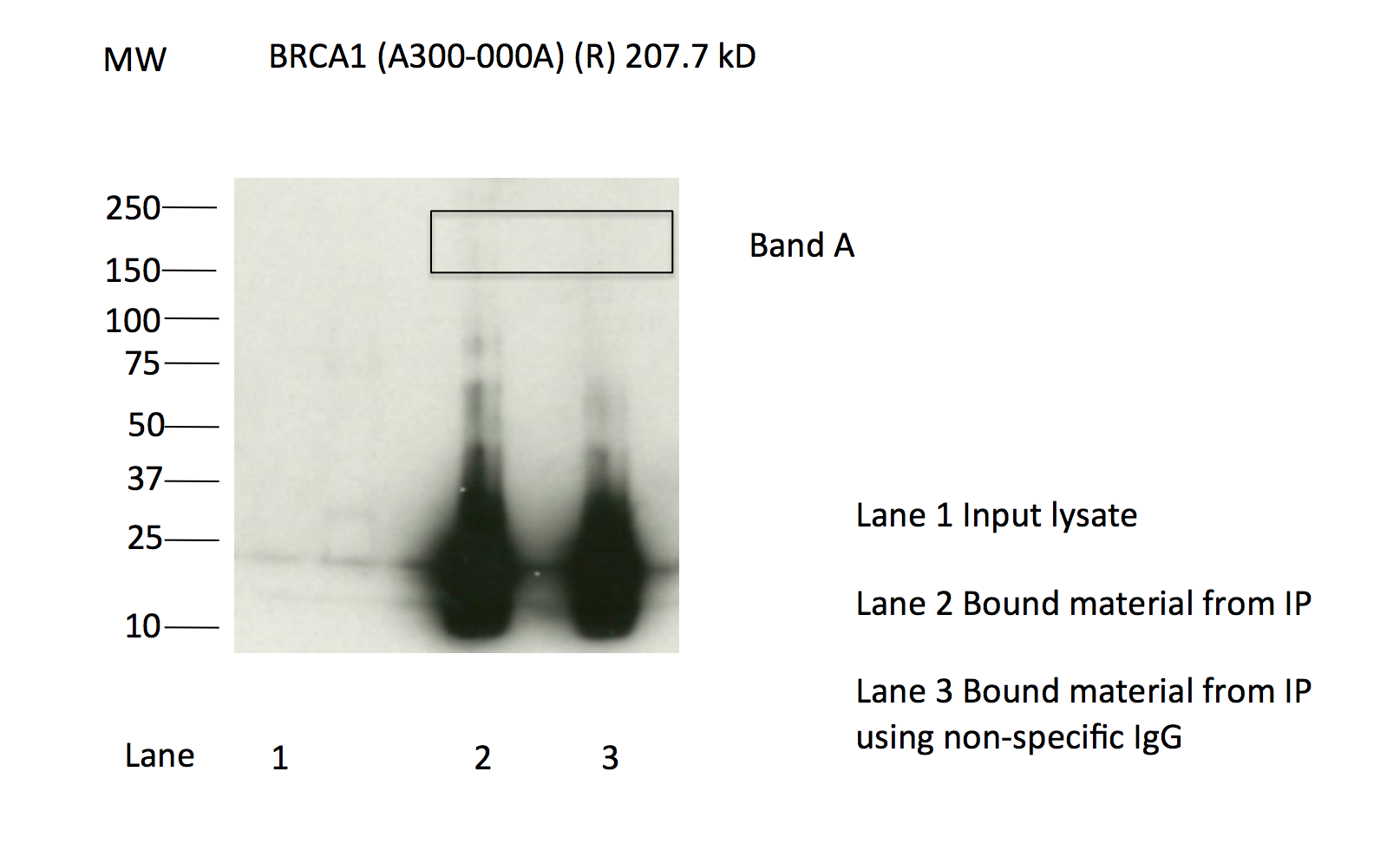
- Immunoprecipitation of BRCA1 from K562 cells using A300-000A. Lane 1: input nuclear lysate, Lane 2: material immunoprecipitated with A300-000A, Lane 3: material immunoprecipitated using control IgG. Bands A was excised from the gel and subject to analysis by mass spectrometry. IP followed by masspectrometry: Briefly, protein was immunoprecipitated from K562 whole cell lysates using A300-000A, and the IP fraction was loaded on a 10% polyacrylamide gel (NuPAGE Bis-Tris Gel) and separated with an Invitrogen NuPAGE electrophoresis system. The gel was silver-stained, gel fragments corresponding to the bands indicated were excised and destained using the SilverSNAP Stain for Mass Spectrometry (Pierce). Then proteins were trypsinized using the in-gel digestion method. Digested proteins were analyzed on an LTQ-Orbitrap (Thermo Scientific) by the nanoLC-ESI-MS/MS technique. Peptides were identified by the SEQUEST algorithm and filtered with a high confidence threshold (Protein false discovery rate < 1%, 2 peptides per protein minimum). P followed by masspectrometry: Briefly, protein was immunoprecipitated from K562 whole cell lysates using A300-000A, and the IP fraction was loaded on a 10% polyacrylamide gel (NuPAGE Bis-Tris Gel) and separated with an Invitrogen NuPAGE electrophoresis system. The gel was silver-stained, gel fragments corresponding to the bands indicated were excised and destained using the SilverSNAP Stain for Mass Spectrometry (Pierce). Then proteins were trypsinized using the in-gel digestion method. Digested proteins were analyzed on an LTQ-Orbitrap (Thermo Scientific) by the nanoLC-ESI-MS/MS technique. Peptides were identified by the SEQUEST algorithm and filtered with a high confidence threshold (Protein false discovery rate < 1%, 2 peptides per protein minimum). Based on these observations, this band is likely due to the presence of immunoprecipitated BRCA1 and A300-000A meets the ENCODE standard for validation by this criterion.
- Submitted by
- Michael Snyder
- Lab
- Michael Snyder, Stanford
- Grant
- U54HG004558
- Download
- human_BRCA1_validation_Snyder.pdf
BRCA1 (Homo sapiens)
K562
compliant
- Caption
- Immunoprecipitation of BRCA1 from K562 cells using A300-000A. Lane 1: input nuclear lysate, Lane 2: material immunoprecipitated with A300-000A, Lane 3: material immunoprecipitated using control IgG. Bands A was excised from the gel and subject to analysis by mass spectrometry. IP followed by masspectrometry: Briefly, protein was immunoprecipitated from K562 whole cell lysates using A300-000A, and the IP fraction was loaded on a 10% polyacrylamide gel (NuPAGE Bis-Tris Gel) and separated with an Invitrogen NuPAGE electrophoresis system.
- Reviewer comment
- No band visible in IP-western but the factor is detected in the cut band by mass-spec.
- Submitted by
- Michael Snyder
- Lab
- Michael Snyder, Stanford
- Grant
- U54HG004558
- Download
- IP + MS for 000AEL.png
BRCA1 (Homo sapiens)
GM12878K562HeLa-S3HepG2
not compliant
- Caption
- Western blots on nuclear lysates from cell lines GM12878 (Lane1), K562 (Lane2), HeLaS3 (Lane3), and HepG2 (Lane4). A band of ~207kD is detected by Western blotting with A300-000A in K562 and HelaS3 nuclear lysates
- Reviewer comment
- Major/marked band not within 20% of expected size
- Submitted by
- Michael Snyder
- Lab
- Michael Snyder, Stanford
- Grant
- U54HG004558
- Download
- WB for 000AEL.png