ENCAB000GBD
Antibody against Homo sapiens ZBTB7B
Homo sapiens
MCF-7, HepG2, K562
characterized to standards
Homo sapiens
any cell type or tissue
partially characterized
Homo sapiens
HEK293T
not characterized to standards
- Status
- released
- Source (vendor)
- Bethyl Labs
- Product ID
- A303-701A
- Lot ID
- 1
- Characterized targets
- ZBTB7B (Homo sapiens)
- Host
- rabbit
- Clonality
- polyclonal
- Purification
- affinity
- Aliases
- michael-snyder:AS-1045
- External resources
Characterizations
ZBTB7B (Homo sapiens)
K562
compliant
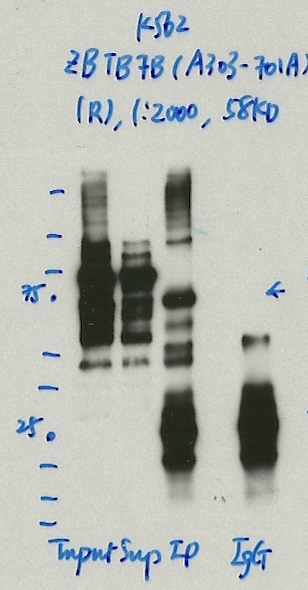
- Caption
- Immunoprecipitation was performed on nuclear extracts from the cell line: K562, using the antibody A303-701A. The blot shows western blot analysis of input, flowthrough, immunoprecipitate and mock immunoprecipitate using IgG.Molecular Weight: 58.027
- Submitted by
- Nathaniel Watson
- Lab
- Michael Snyder, Stanford
- Grant
- U54HG006996
- Download
- 1108_11_ZBTB7B_A303-701A.jpg
ZBTB7B (Homo sapiens)
HepG2
compliant
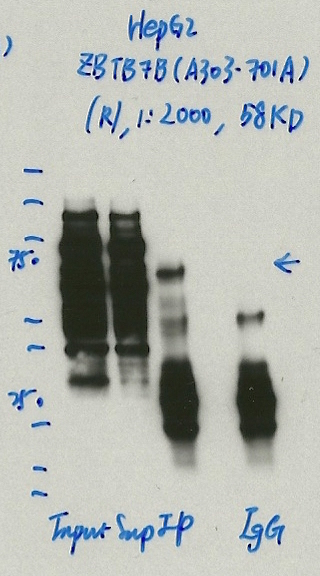
- Caption
- Immunoprecipitation was performed on nuclear extracts from the cell line: HepG2, using the antibody A303-701A. The blot shows western blot analysis of input, flowthrough, immunoprecipitate and mock immunoprecipitate using IgG.Molecular Weight: 58.027
- Submitted by
- Nathaniel Watson
- Lab
- Michael Snyder, Stanford
- Grant
- U54HG006996
- Download
- 1108_12_ZBTB7B_A303-701A.jpg
ZBTB7B (Homo sapiens)
HEK293T
not compliant
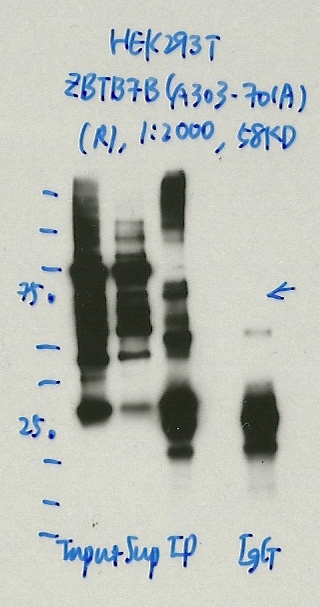
- Caption
- Immunoprecipitation was performed on nuclear extracts from the cell line: HEK293T, using the antibody A303-701A. The blot shows western blot analysis of input, flowthrough, immunoprecipitate and mock immunoprecipitate using IgG.Molecular Weight: 58.027
- Submitted by
- Nathaniel Watson
- Lab
- Michael Snyder, Stanford
- Grant
- U54HG006996
- Download
- 1112_11_ZBTB7B_A303-701A.jpg
ZBTB7B (Homo sapiens)
Method: motif enrichment
compliant
- Caption
- The motif for target ZBTB7B is represented by the attached position weight matrix (PWM) derived from ENCFF070WUF. Motif enrichment analysis was done by Dr. Zhizhuo Zhang (Broad Institute, Kellis Lab) using known motifs (http://compbio.mit.edu/encode-motifs/) and previously published ChIP-seq data (http://www.broadinstitute.org/~zzhang/motifpipeline/data/TrainSetInfo.txt). The accept probability score of the given transcription factor was calculated using a Bayesian approach. This analysis also includes three motif enrichment scores, computed by overlapping the motif instances with the given ChIP-seq peak locations, as well as an enrichment rank. For more information on the underlying statistical methods, please see the attached document. Accept probability score: 0.681678279217 Global enrichment Z-score: -0.152665158197 Positional bias Z-score: 1.59322956416 Peak rank bias Z-score: 1.66599203246 4 Enrichment rank: 1.0
- Submitted by
- Jessika Adrian
- Lab
- Michael Snyder, Stanford
- Grant
- U54HG006996
ZBTB7B (Homo sapiens)
MCF-7
not compliant
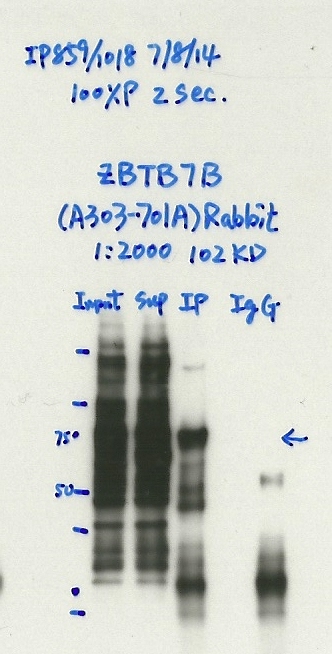
- Caption
- Immunoprecipitation was performed on nuclear extracts from the cell line: MCF-7, using the antibody A303-701A. The blot shows western blot analysis of input, flowthrough, immunoprecipitate and mock immunoprecipitate using IgG.Molecular Weight: 58.027
- Reviewer comment
- Gel says 102 KD, Uniprot says 58 KD. Still not compliant, not 50% of signal
- Submitted by
- Denis Salins
- Lab
- Michael Snyder, Stanford
- Grant
- U54HG006996
- Download
- Expt 859_5-ZBTB7B0002.jpg
ZBTB7B (Homo sapiens)
not submitted for review by lab
- Caption
- Immunoprecipitation was performed on nuclear extracts from the cell line: GM12878, using the antibody A303-701A. The blot shows western blot analysis of input, flowthrough, immunoprecipitate and mock immunoprecipitate using IgG.Molecular Weight: 58.027
- Submitted by
- Nathaniel Watson
- Lab
- Michael Snyder, Stanford
- Grant
- U54HG006996
- Download
- expt_1107_6-ZBTB7B-A303-701A.jpg
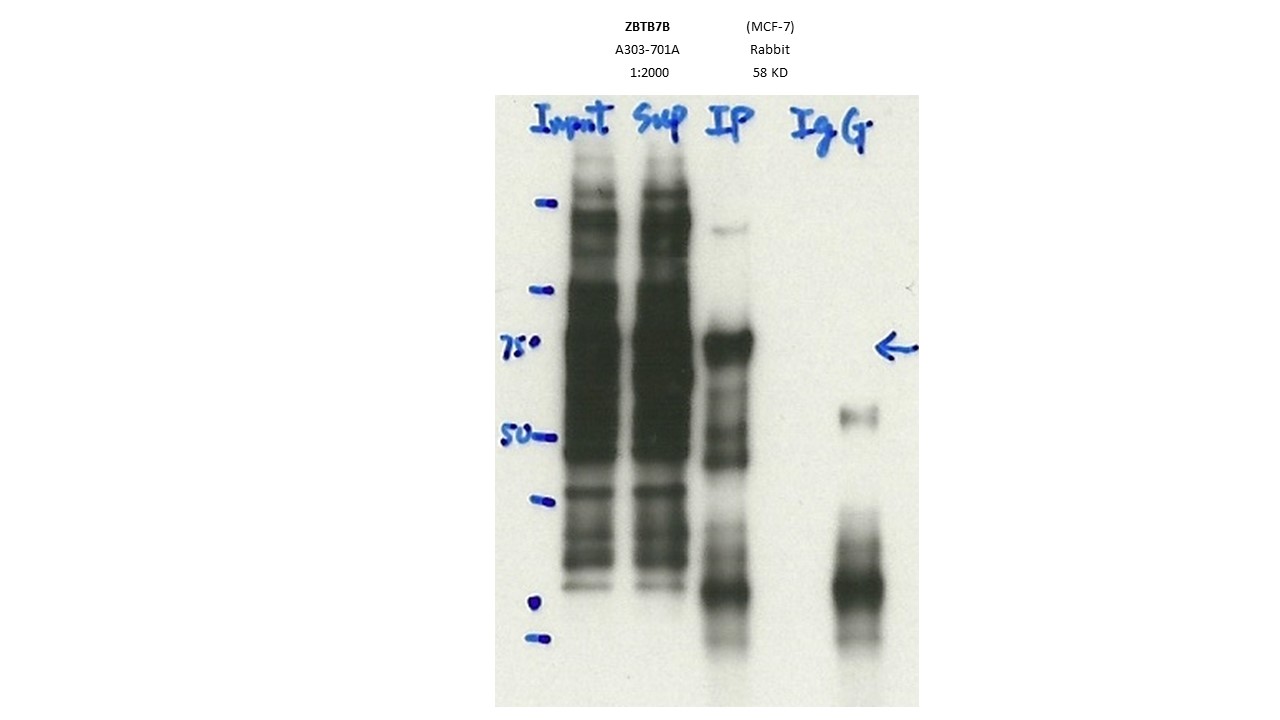
ZBTB7B (Homo sapiens)
MCF-7
compliant
- Caption
- Immunoprecipitation was performed on nuclear extracts from the cell line: MCF-7, using the antibody A303-701A. The image shows western blot analysis of input, flowthrough, immunoprecipitate, and mock immunoprecipitate using IgG. Target molecular weight: 58.027.
- Submitted by
- Nathaniel Watson
- Lab
- Michael Snyder, Stanford
- Grant
- U54HG006996
- Download
- IP1018_ZBTB7B_.jpg