ENCAB000ANT
Antibody against Homo sapiens YY1
Homo sapiens
HeLa-S3, GM12878, K562
characterized to standards
Homo sapiens
HepG2, liver
characterized to standards with exemption
- Status
- released
- Source (vendor)
- Santa Cruz Biotech
- Product ID
- sc-281
- Lot ID
- B1010
- Characterized targets
- YY1 (Homo sapiens)
- Lot ID aliases
- F2810
- Host
- rabbit
- Clonality
- polyclonal
- Isotype
- IgG
- Antigen description
- Epitope mapping at the C-terminus of YY1 of human origin.
- External resources
Characterizations
YY1 (Homo sapiens)
Method: motif enrichment
compliant
- Caption
- The motif for target YY1 is represented by the attached position weight matrix (PWM) derived from files ENCFF933CGK and ENCFF959SQX. Motif enrichment analysis was done by Dr. Zhizhuo Zhang (Broad Institute, Kellis Lab) using known motifs (http://compbio.mit.edu/encode-motifs/) and previously published ChIP-seq data (http://www.broadinstitute.org/~zzhang/motifpipeline/data/TrainSetInfo.txt). The accept probability score of the given transcription factor was calculated using a Bayesian approach. This analysis also includes three motif enrichment scores, computed by overlapping the motif instances with the given ChIP-seq peak locations. For more information on the underlying statistical methods, please see the attached document. From ENCFF933CGK: Accept probability score: 0.940177543827, Global enrichment Z-score: 5.00869333435, Positional bias Z-score: 5.25834816581, Peak rank bias Z-score: 10.5448880879. From ENCFF959SQX: Accept probability score: 0.940177543827, Global enrichment Z-score: 4.32978569726, Positional bias Z-score: 5.25834816581, Peak rank bias Z-score: 9.33391043935, Enrichment rank: 1.0..
- Submitted by
- Aditi Narayanan
- Lab
- Richard Myers, HAIB
- Grant
- U54HG006998
- Download
- ENCAB000ANT.pdf
YY1 (Homo sapiens)
HepG2HeLa-S3GM12878K562
compliant
- Caption
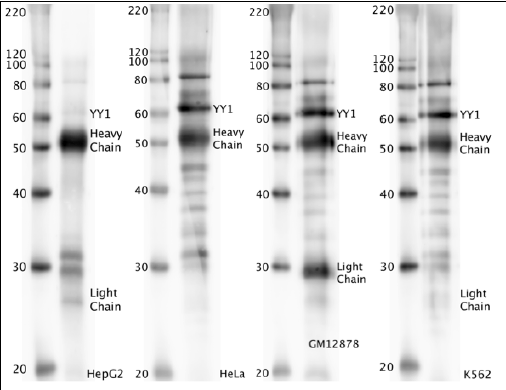
- Western blot protocol: Whole cell lysate was immunoprecipitated using primary antibody, and the IP fraction was loaded on a 12% acrylamide gel and separated with a Bio-Rad PROTEAN II xi system. After separation, the samples were transferred to a nitrocellulose membrane with an Invitrogen iBlot system. Blotting with primary (same as that used for IP) and secondary HRP-conjugated antibodies was performed on an Invitrogen BenchPro 4100 system. Visualization was achieved using SuperSignal West Femto solution (Thermo Scientific). Expected size ~44.7 kDa. Results: Band of expected size visualized, representing strongest signal in the lane. Figure legend: IP-western with sc-281 in whole cell lysate (WCL) of HepG2, HeLa, GM12878 and K562; PM=protein marker. YY1 bands are indicated.
- Reviewer comment
- Reviewed by Dr. Peggy Farnham, who wishes to exempt the HepG2 lane from standards
- Submitted by
- Richard Myers
- Lab
- Richard Myers, HAIB
- Grant
- U54HG004576
YY1 (Homo sapiens)
liver
exempt from standards
- Caption
- The ENCODE Binding Working Group finds for some valuable tissues that recreating a primary on well characterized antibodies is not cost effective. Therefore, they allow exemption from standards for these tissues.
- Submitter comment
- The lab is asking for an exemption for liver cells due to the lack of resource to make a primary characterization for them
- Reviewer comment
- Exempted by the Feb 29, 2016 antibody review panel
- Submitted by
- Richard Myers
- Lab
- Richard Myers, HAIB
- Grant
- U54HG006998
- Download
- No_tissue.png
YY1 (Homo sapiens)
not reviewed
- Caption
- IP followed by mass spectrometry: Briefly, K562 whole cell lysates were immunoprecipitated using primary antibody, and the IP fraction was loaded on a 12% acrylamide gel and separated with a Bio-Rad PROTEAN II xi system. Gel was stained with Coomassie Blue in order to visualize marker bands. Gel fragments corresponding to the bands indicated above in the western blot image were excised and sent to the University of Alabama at Birmingham Cancer Center Mass Spectrometry/Proteomics Shared Facility. There the samples were run on an LTQ XL Linear Ion Trap Mass Spectrometer by LC-ESI-MS/MS. Peptides were identified using SEQUEST tandem mass spectra analysis, with probability based matching at p < 0.05. As per ENCODE data standards, all SEQUEST results are attached (ENCODE_HAIB_YY1_sc281_09122011_MassSpec.pdf), including common contaminants. Target protein is listed as hit 17 in ~60 kDa band.
- Submitted by
- Richard Myers
- Lab
- Richard Myers, HAIB
- Grant
- U54HG004576
- Documents