ENCAB000AKG
Antibody against Homo sapiens RAD21, Mus musculus RAD21
Homo sapiens
K562, GM12878, HeLa-S3, HepG2
characterized to standards
Mus musculus
CH12.LX, MEL
characterized to standards
Homo sapiens
liver, neural cell
characterized to standards with exemption
Homo sapiens
MCF-7
not characterized to standards
- Status
- released
- Source (vendor)
- Abcam
- Product ID
- ab992
- Lot ID
- 940739
- Characterized targets
- RAD21 (Homo sapiens), RAD21 (Mus musculus)
- Lot ID aliases
- 891751, 734371
- Host
- rabbit
- Clonality
- polyclonal
- Purification
- affinity
- Antigen description
- Synthetic peptide (Human) conjugated to KLH - which represents a portion of human Rad21 encoded within exon 14
- External resources
Characterizations
RAD21 (Homo sapiens)
neural cell
exempt from standards
- Caption
- The ENCODE antibody standards document exempts some valuable and/or limiting samples from primary characterizations for well-characterized antibodies. They are given exemptions so that the samples can be conserved to carry out the downstream experiments.
- Submitter comment
- We do not have any of the H1-derived neurons that were distributed to the consortium for analysis in ENCODE2 left.
- Reviewer comment
- This cell type was exempted from primary characterization of antibody ENCAB000AIT by the ENCODE antibody review panel on October 17, 2016
- Submitted by
- Jessika Adrian
- Lab
- Michael Snyder, Stanford
- Grant
- U54HG006996
- Download
- H1neuron_primary.png
RAD21 (Homo sapiens)
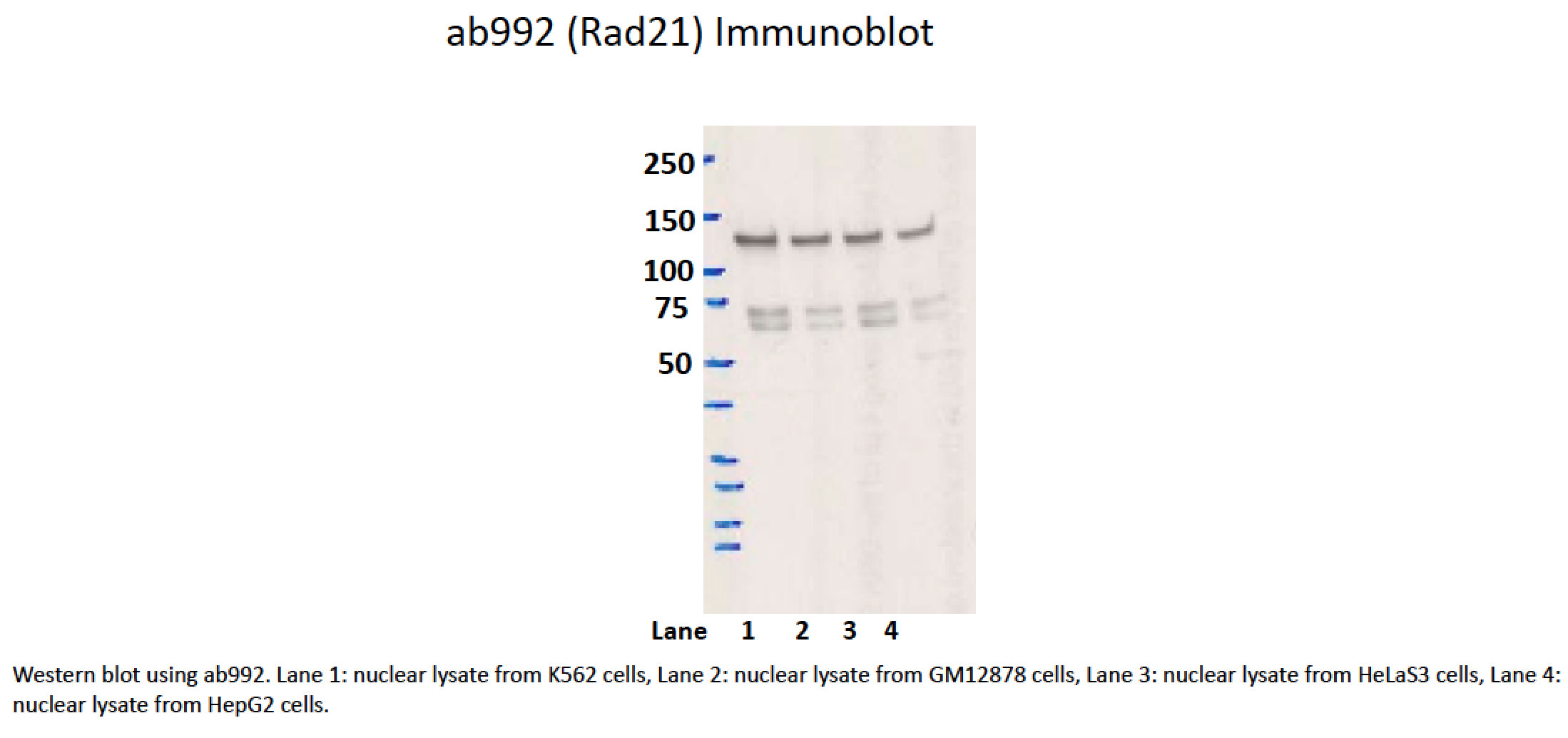
K562GM12878HeLa-S3HepG2
compliant
- Caption
- While the predicted size of Rad21 is ~72kD, according to product characterization literature (found at manufacturer's website), the bands at ~70kD are of unknown origin while a band of ~140kD, similar to the one observed here, is identified as Rad21. In support of this, siRNAs targeting Rad21 dramatically reduce levels of the 140kD band, while levels of the 70kD band are unaffected (see Figure 2 of this document). 1. Hyperphosphorylation (ref. PMID 11073952 & many others) https://www.ncbi.nlm.nih.gov/gene/5885 2. Dimerisation as part of cohesin complex (whose other members we can detect in the 130kDa band, refs. PMIDs 23874961 & 19075111)
- Reviewer comment
- The lab has provided in the literature two possible explanations for the difference in size of the bands. This is confirmed by the siRNA knockdown in the other characterization associated with this antibody.
- Submitted by
- Michael Snyder
- Lab
- Michael Snyder, Stanford
- Grant
- U54HG004558
RAD21 (Mus musculus)
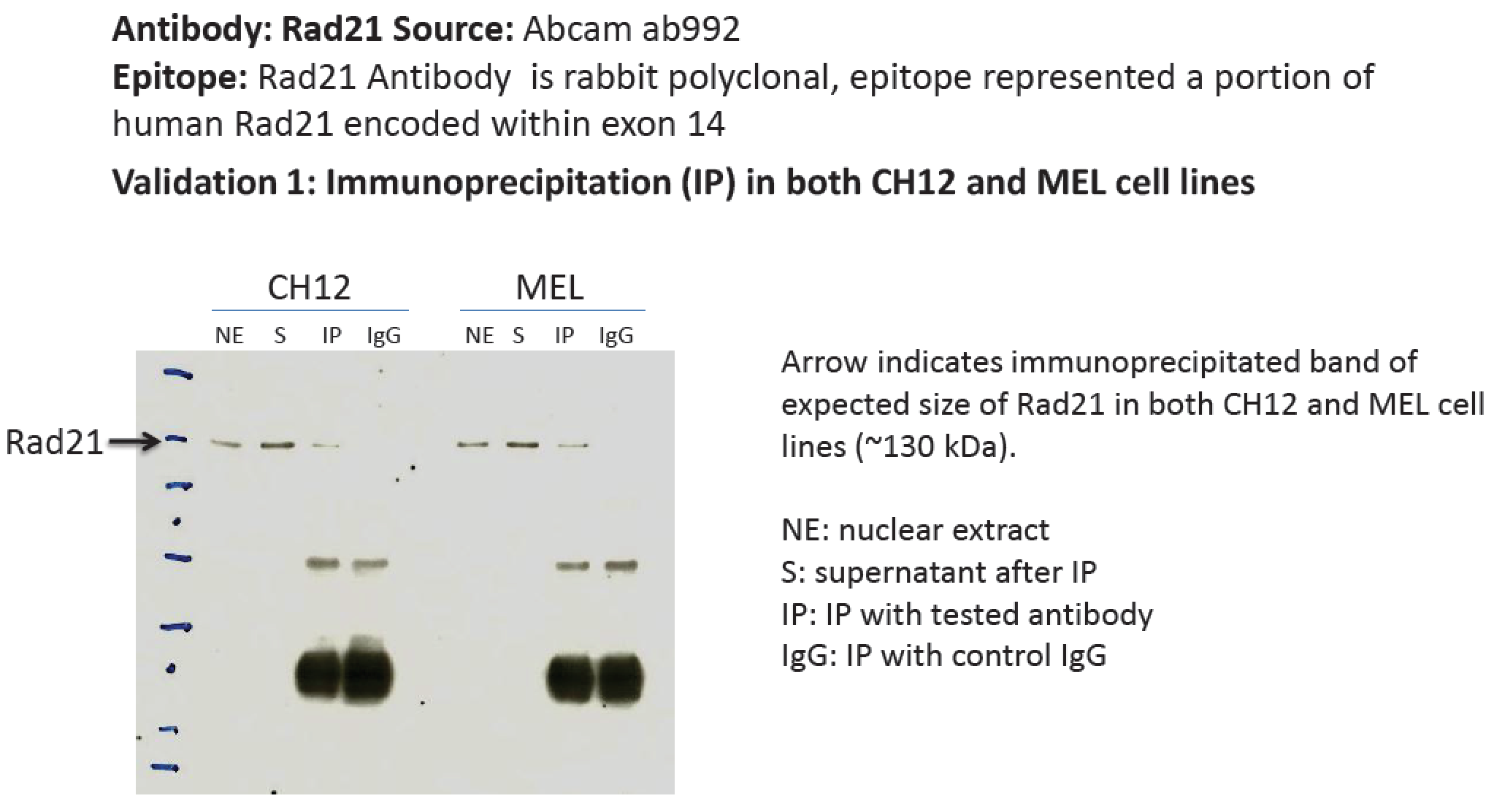
CH12.LXMEL
compliant
- Caption
- A band of ~130kD was immunoprecipitated from CH12 and MEL nuclear extracts using ab992. This antibody has been validated for human cell lines by Immunoprecipitation and siRNA knockdown.
- Submitter comment
- While the predicted size of Rad21 is ~72kD, according to product characterization literature (found at manufacturer's website), the bands at ~70kD are of unknown origin while a band of ~140kD, similar to the one observed here, is identified as Rad21. In support of this, siRNAs targeting Rad21 dramatically reduce levels of the 140kD band, while levels of the 70kD band are unaffected (see Figure 2 of this document). 1. Hyperphosphorylation (ref. PMID 11073952 & many others) https://www.ncbi.nlm.nih.gov/gene/5885 2. Dimerisation as part of cohesin complex (whose other members we can detect in the 130kDa band, refs. PMIDs 23874961 & 19075111)
- Reviewer comment
- According to Uniprot, Rad21 is ~71-73 kDa, not ~130. Also not 50% of overall signal
- Submitted by
- Michael Snyder
- Lab
- Michael Snyder, Stanford
- Grant
- RC2HG005602
RAD21 (Homo sapiens)
liver
exempt from standards
- Caption
- The ENCODE Binding Working Group finds for some valuable tissues that recreating a primary on well characterized antibodies is not cost effective. Therefore, they allow exemption from standards for these tissues.
- Submitter comment
- The lab is asking for an exemption for liver cells due to the lack of resource to make a primary characterization for them
- Reviewer comment
- Exempted by the Feb 29, 2016 antibody review panel
- Submitted by
- Richard Myers
- Lab
- Richard Myers, HAIB
- Grant
- U54HG006998
- Download
- No_tissue.png
RAD21 (Homo sapiens)
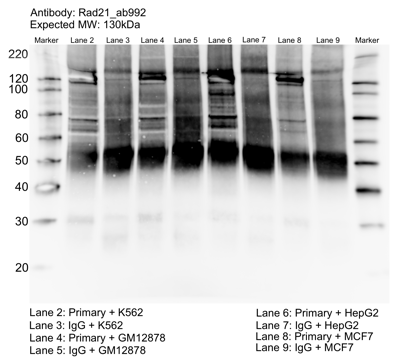
K562GM12878HepG2MCF-7
not compliant
- Caption
- Whole cell lysates of K562, MCF7, GM12878 and HepG2 were immunoprecipitated using the primary antibody (Abcam; ab992). The IP fraction was separated on a 12% acrylamide gel with the Bio-Rad PROTEAN II xi system. After separation, the samples were transferred to a nitrocellulose membrane with an Invitrogen iBlot system. The membrane was probed with the primary antibody (same as that used for IP) and a secondary HRP-conjugated antibody. The resulting bands were visualized with SuperSignal West Femto Solution (Thermo Scientific). Protein Marker (PM) is labeled in kDa. The approximate size of RAD21 is ~130 kDa.
- Submitted by
- Mark Mackiewicz
- Lab
- Richard Myers, HAIB
- Grant
- U54HG004576
- Download
- Rad21_ab992_092916_L.png
RAD21 (Homo sapiens)
compliant
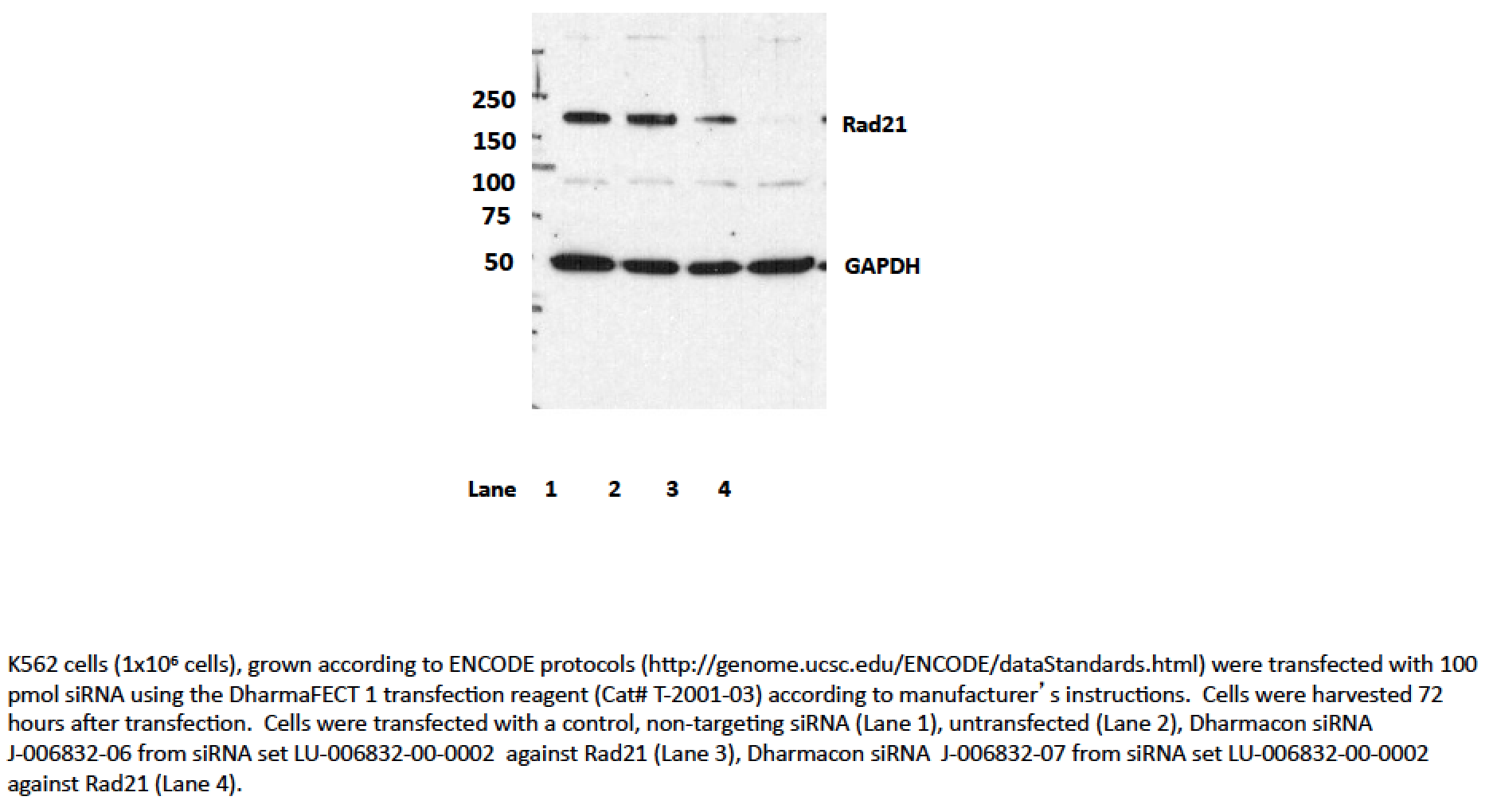
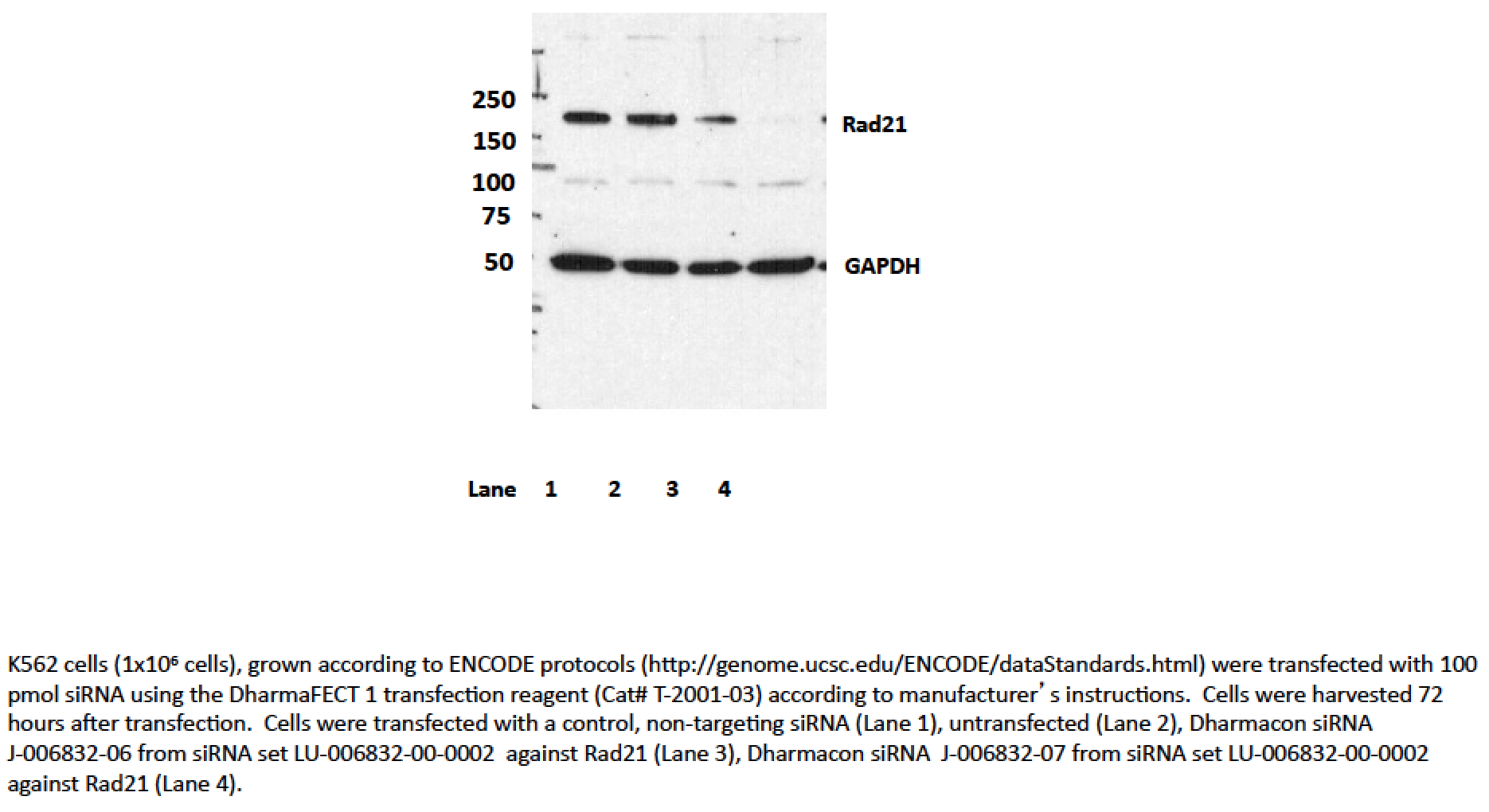
- Caption
- Transfection with siRNA 7, and to a lesser extent, siRNA 6 significantly reduces levels of the ~145kD band seen by Western blotting with ab992 in all cell lines, while levels of the lower abundance ~70kD band are unaffected. As a loading control, blots were also probed with an antibody to GAPDH, which was also unaffected by siRNA treatment (indicated by arrow). Based on these observations, ab992 is validated by this ENCODE criterion.We also note that an additional validation assay, ChIP with an antibody against SMC3 (a member of the same protein complex as Rad21) has been carried out and very high levels of agreement are observed between ChIP experiments. Data is available in the validation document for ENCODE antibody ab9263.
- Submitter comment
- According to our rules, if siRNA treatment results in knockdown of the major band (regardless if it is the size expected from the translation of the coding sequence), then the siRNA has “rescued” the antibody and it passes.
- Submitted by
- Michael Snyder
- Lab
- Michael Snyder, Stanford
- Grant
- U54HG004558
RAD21 (Mus musculus)
not reviewed
- Caption
- Transfection with siRNA 7, and to a lesser extent, siRNA 6 significantly reduces levels of the ~145kD band seen by Western blotting with ab992 in all cell lines, while levels of the lower abundance ~70kD band are unaffected. As a loading control, blots were also probed with an antibody to GAPDH, which was also unaffected by siRNA treatment (indicated by arrow). Based on these observations, ab992 is validated by this ENCODE criterion.We also note that an additional validation assay, ChIP with an antibody against SMC3 (a member of the same protein complex as Rad21) has been carried out and very high levels of agreement are observed between ChIP experiments. Data is available in the validation document for ENCODE antibody ab9263.
- Submitted by
- Michael Snyder
- Lab
- Michael Snyder, Stanford
- Grant
- RC2HG005602
- Documents