ENCAB000AHG
Antibody against Homo sapiens HCFC1, Mus musculus HCFC1
Homo sapiens
K562, GM12878, HepG2
characterized to standards
Homo sapiens
MCF-7, HeLa-S3
characterized to standards with exemption
Homo sapiens
any cell type or tissue
partially characterized
Mus musculus
any cell type or tissue
partially characterized
- Status
- released
- Source (vendor)
- Novus
- Product ID
- NB100-68209
- Lot ID
- A1
- Characterized targets
- HCFC1 (Homo sapiens), HCFC1 (Mus musculus)
- Host
- rabbit
- Clonality
- polyclonal
- Antigen description
- The epitope recognized by this antibody maps to a region between residue 1700 and 1750 of human host cell factor C1 (VP16-accessory protein).
- External resources
Characterizations
HCFC1 (Homo sapiens)
GM12878
compliant
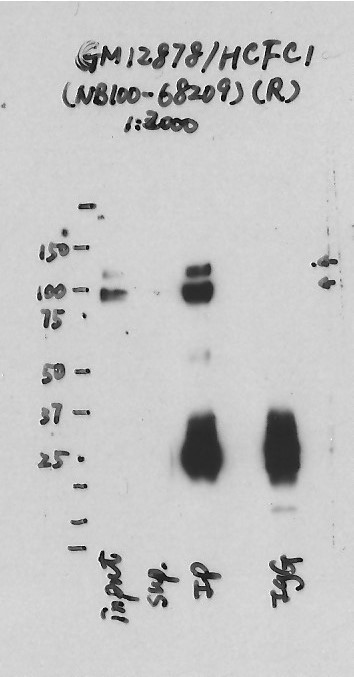
- Caption
- Immunoprecipitation was performed on nuclear extracts from the cell line: GM12878, using the antibody NB100-68209. The blot shows western blot analysis of input, flowthrough, immunoprecipitate and mock immunoprecipitate using IgG.Molecular Weight: 208.732
- Submitted by
- Nathaniel Watson
- Lab
- Michael Snyder, Stanford
- Grant
- U54HG006996
- Download
- GM12878-HCFC1.jpg
HCFC1 (Homo sapiens)
HepG2
compliant
- Caption
- Immunoprecipitation was performed on nuclear extracts from the cell line: HepG2, using the antibody NB100-68209. The blot shows western blot analysis of input, flowthrough, immunoprecipitate and mock immunoprecipitate using IgG.Molecular Weight: 208.732
- Submitted by
- Nathaniel Watson
- Lab
- Michael Snyder, Stanford
- Grant
- U54HG006996
- Download
- HCFC1.JPG
HCFC1 (Homo sapiens)
Method: immunoprecipitation followed by mass spectrometry
compliant
- Caption
- IP followed by mass spectrometry: Briefly, protein was immunoprecipitated from K562 whole cell lysates using NB100-68209, and the IP fraction was loaded on a 10% polyacrylamide gel (NuPAGE Bis-Tris Gel) and separated with an Invitrogen NuPAGE electrophoresis system. The gel was silver-stained, gel fragments corresponding to the bands indicated were excised and destained using the SilverSNAP Stain for Mass Spectrometry (Pierce). Then proteins were trypsinized using the in-gel digestion method. Digested proteins were analyzed on an LTQ-Orbitrap (Thermo Scientific) by the nanoLC-ESI-MS/MS technique. Peptides were identified by the SEQUEST algorithm and filtered with a high confidence threshold (Protein false discovery rate < 1%, 2 peptides per protein minimum). We report 29 different proteins identified in band A and B. Of the specifically immunoprecipitated proteins, HCFC1 is the most abundant protein in both band A as well as band B.
- Submitted by
- Kathrina Onate
- Lab
- Michael Snyder, Stanford
- Grant
- U54HG004558
- Download
- HCFC1_final_AHG Sheet1.pdf
HCFC1 (Homo sapiens)
Method: immunoprecipitation followed by mass spectrometry
not reviewed
- Submitted by
- Michael Snyder
- Lab
- Michael Snyder, Stanford
- Grant
- U54HG004558
HCFC1 (Homo sapiens)
Method: immunoprecipitation
not reviewed
- Caption
- ChIPseq was preformed in Hela cells stably transfected with a plasmid expressing an HA tagged E2F1 protein. Two different biological replicates of HeLa cells were used; one sample was ChIPPed using the HA antibody and the other sample was ChIPPed useing the E2F1 antibody. The antibodies showed very similar ChIPseq profiles: 99% of the top 40% of targets identified with the HA antubody are in the targets identified using the E2F1 antibody and 97.5% of the top 40% of targets identified using the E2F1 antibody are in the targets identified using the HA antibody.
- Submitted by
- Michael Snyder
- Lab
- Michael Snyder, Stanford
- Grant
- U54HG004558
HCFC1 (Homo sapiens)
K562
exempt from standards
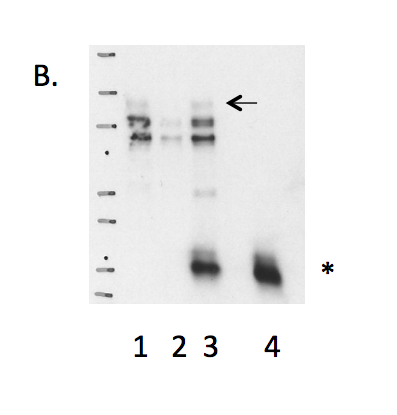
- Caption
- B. Immunoprecipitation was performed on nuclear lysates from K562 cells using antibody NB100-68209 against HCFC1. Lane1: Nuclear lysate. Lane 2: Unbound material from immunoprecipitation with NB100-68209. Lane 3: Bound material from immunoprecipitation with NB100-68209. Lane 4: Bound material from control immunoprecipitation with rabbit IgG. Arrow indicates band of expected size (208 kD) that is enriched in the specifically immunoprecipitated fraction. Smaller band detected in the IP is possibly degradation product as indicated by the Mass Spec analysis. Band indicated by * in K562 immunoprecipitate is IgG light chains.
- Submitter comment
- The protein undergoes proteolytic cleavage.
- Reviewer comment
- Observed bands are outside of 20% of the expected size (~208kD). However, it is known to undergo proteolytic cleavage into a smaller final form (http://www.genecards.org/cgi-bin/carddisp.pl?gene=HCFC1) and the shown bands is consistent with the forms detected by the vendor (http://www.novusbio.com/Host-Cell-Factor-1-HCFC1-Antibody_NB100-68209.html#dsTab1).
- Submitted by
- Kathrina Onate
- Lab
- Michael Snyder, Stanford
- Grant
- U54HG004558
- Download
- IP Snyder AHG.png
HCFC1 (Homo sapiens)
K562
compliant
- Caption
- Immunoprecipitation of HCFC1 from K562 cells using NB100-68209. Lane 1: input nuclear lysate, Lane 2: material immunoprecipitated with NB100-68209, Lane 3: material immunoprecipitated using control IgG. Bands A and B were excised from the gel and subject to analysis by mass spectrometry.
- Submitted by
- Kathrina Onate
- Lab
- Michael Snyder, Stanford
- Grant
- U54HG004558
- Download
- IP-MS_HCFC1:K562 Snyder AHG.png
HCFC1 (Homo sapiens)
MCF-7
exempt from standards
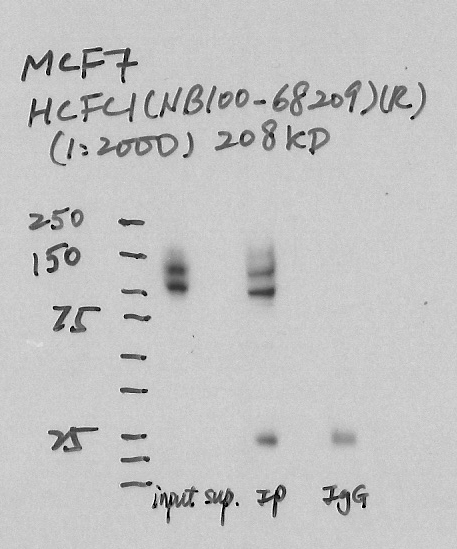
- Caption
- Immunoprecipitation was performed on nuclear extracts from the cell line: MCF-7, using the antibody NB100-68209. The blot shows western blot analysis of input, flowthrough, immunoprecipitate and mock immunoprecipitate using IgG.
- Submitter comment
- Protein undergoes proteolytic cleavage.
- Reviewer comment
- Major band not within 20% of the expected size of the protein, no lane labels, no arrow indicating expected size. It appears that only the proteolytic fragments were detected: http://www.novusbio.com/Host-Cell-Factor-1-HCFC1-Antibody_NB100-68209.html#dsTab1
- Submitted by
- Denis Salins
- Lab
- Michael Snyder, Stanford
- Grant
- U54HG006996
HCFC1 (Mus musculus)
Method: immunoprecipitation followed by mass spectrometry
not reviewed
- Submitted by
- Michael Snyder
- Lab
- Michael Snyder, Stanford
- Grant
- RC2HG005602
- Download
- mouse_HCFC1_validation_Snyder.pdf
HCFC1 (Mus musculus)
Method: immunoprecipitation
not reviewed
- Submitted by
- Michael Snyder
- Lab
- Michael Snyder, Stanford
- Grant
- RC2HG005602
- Download
- mouse_HCFC1_validation_Snyder.pdf
HCFC1 (Homo sapiens)
GM12878K562HeLa-S3HepG2
exempt from standards
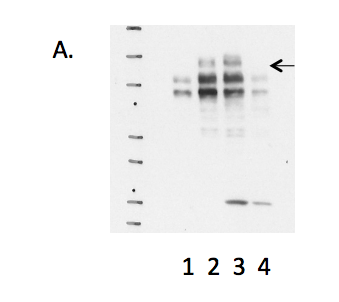
- Caption
- A. Western blots on nuclear lysates from cell lines GM12878 (Lane1), K562 (Lane2), HeLaS3 (Lane3), and HepG2 (Lane4). The expected band size is ~208 kD (indicated by arrow).
- Submitter comment
- Protein undergoes proteolytic cleavage; desired band is marked with arrow.
- Reviewer comment
- Observed bands are outside of 20% of the expected size (~208kD). However, it is known to undergo proteolytic cleavage into a smaller final form (http://www.genecards.org/cgi-bin/carddisp.pl?gene=HCFC1) and the shown bands is consistent with the forms detected by the vendor (http://www.novusbio.com/Host-Cell-Factor-1-HCFC1-Antibody_NB100-68209.html#dsTab1).
- Submitted by
- Kathrina Onate
- Lab
- Michael Snyder, Stanford
- Grant
- U54HG004558
- Download
- WB Snyder AHG.png