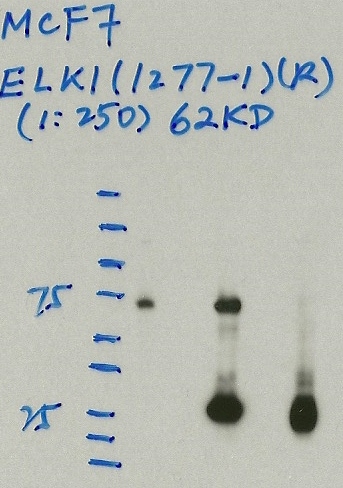ENCAB000AGB
Antibody against Homo sapiens ELK1
Homo sapiens
MCF-7, K562, IMR-90, A549
characterized to standards
Homo sapiens
any cell type or tissue
partially characterized
- Status
- released
- Source (vendor)
- Epitomics
- Product ID
- 1277-1
- Lot ID
- YG030402C
- Characterized targets
- ELK1 (Homo sapiens)
- Host
- rabbit
- Clonality
- polyclonal
- Purification
- affinity
- Antigen description
- Synthetic peptide corresponding to residues in C-terminus of human ELK-1 was used as immunogen. The antibody does not cross-react with other ETS family members.
- External resources
Characterizations
ELK1 (Homo sapiens)
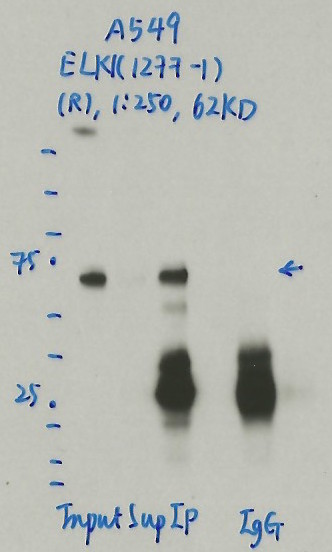
A549
compliant
- Caption
- Immunoprecipitation was performed on nuclear extracts from the cell line: A549, using the antibody 1277-1. The blot shows western blot analysis of input, flowthrough, immunoprecipitate and mock immunoprecipitate using IgG.Molecular Weight: 62.0
- Submitted by
- Nathaniel Watson
- Lab
- Michael Snyder, Stanford
- Grant
- U54HG006996
- Download
- 1125_01_ELK1_1277-1.jpg
ELK1 (Homo sapiens)
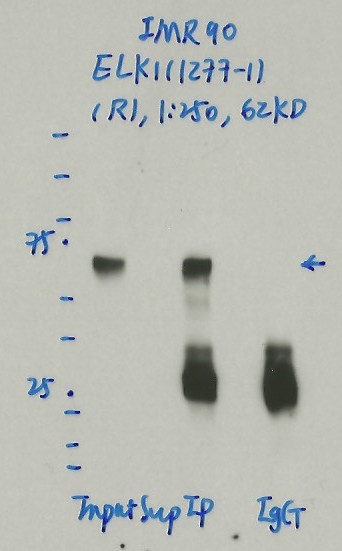
IMR-90IMR-90
compliant
- Caption
- Immunoprecipitation was performed on nuclear extracts from the cell line: IMR90, using the antibody 1277-1. The blot shows western blot analysis of input, flowthrough, immunoprecipitate and mock immunoprecipitate using IgG.Molecular Weight: 62.0
- Submitted by
- Nathaniel Watson
- Lab
- Michael Snyder, Stanford
- Grant
- U54HG006996
- Download
- 1125_02_ELK1_1277-1.jpg
ELK1 (Homo sapiens)
MCF-7
compliant
- Caption
- Immunoprecipitation was performed on nuclear extracts from the cell line: MCF-7, using the antibody 1277-1. The blot shows western blot analysis of input, flowthrough, immunoprecipitate and mock immunoprecipitate using IgG.
- Submitted by
- Denis Salins
- Lab
- Michael Snyder, Stanford
- Grant
- U54HG006996
- Download
- 811 1_ ELK1.jpg
ELK1 (Homo sapiens)
Method: immunoprecipitation followed by mass spectrometry
compliant
- Caption
- IP followed by mass spectrometry: Briefly, protein was immunoprecipitated from K562 whole cell lysates using 1277-1, and the IP fraction was loaded on a 10% polyacrylamide gel (NuPAGE Bis-Tris Gel) and separated with an Invitrogen NuPAGE electrophoresis system. The gel was silver-stained, gel fragments corresponding to the bands indicated were excised and destained using the SilverSNAP Stain for Mass Spectrometry (Pierce). Then proteins were trypsinized using the in-gel digestion method. Digested proteins were analyzed on an LTQ-Orbitrap (Thermo Scientific) by the nanoLC-ESI-MS/MS technique. Peptides were identified by the SEQUEST algorithm and filtered with a high confidence threshold (Protein false discovery rate < 1%, 2 peptides per protein minimum).
- Submitted by
- Kathrina Onate
- Lab
- Michael Snyder, Stanford
- Grant
- U54HG004558
- Download
- ELK1_final_AGB Sheet1.pdf
ELK1 (Homo sapiens)
Method: motif enrichment
compliant
- Caption
- The motif for target ELK1 is represented by the attached position weight matrix (PWM) derived from ENCFF732WQJ. Motif enrichment analysis was done by Dr. Zhizhuo Zhang (Broad Institute, Kellis Lab) using known motifs (http://compbio.mit.edu/encode-motifs/) and previously published ChIP-seq data (http://www.broadinstitute.org/~zzhang/motifpipeline/data/TrainSetInfo.txt). The accept probability score of the given transcription factor was calculated using a Bayesian approach. This analysis also includes three motif enrichment scores, computed by overlapping the motif instances with the given ChIP-seq peak locations, as well as an enrichment rank. For more information on the underlying statistical methods, please see the attached document. Accept probability score: 0.857103478 Global enrichment Z-score: 3.305428989 Positional bias Z-score: 3.365156281 Peak rank bias Z-score: 4.004685043 Enrichment rank: 3.0
- Submitted by
- Kathrina Onate
- Lab
- Michael Snyder, Stanford
- Grant
- U54HG006996
ELK1 (Homo sapiens)
Method: immunoprecipitation followed by mass spectrometry
not reviewed
- Caption
- IP followed by mass spectrometry: Briefly, protein was immunoprecipitated from K562 whole cell lysates using 1277-1, and the IP fraction was loaded on a 10% polyacrylamide gel (NuPAGE Bis-Tris Gel) and separated with an Invitrogen NuPAGE electrophoresis system. The gel was silver-stained, gel fragments corresponding to the bands indicated were excised and destained using the SilverSNAP Stain for Mass Spectrometry (Pierce). Then proteins were trypsinized using the in-gel digestion method. Digested proteins were analyzed on an LTQ-Orbitrap (Thermo Scientific) by the nanoLC-ESI-MS/MS technique. Peptides were identified by the SEQUEST algorithm and filtered with a high confidence threshold (Protein false discovery rate < 1%, 2 peptides per protein minimum).
- Submitted by
- Michael Snyder
- Lab
- Michael Snyder, Stanford
- Grant
- U54HG004558
ELK1 (Homo sapiens)
Method: immunoprecipitation
not reviewed
- Caption
- We observe one major band of ~65kD in all cell lines assayed (Panel A). This band is immunoprecipitated from K562 cells at very high efficiency and is absent in an IgG control immunoprecipitation. (Panel B and second validation assay). The major band migrates somewhat slower than predicted for ELK1 (~44kD), but this is generally observed with ELK1 (see product literature for this and other commercially available antibodies). Moreover, this band was confirmed to contain ELK1 by mass spectrometry (see second validation assay). Therefore, 1277-1 meets this criterion for validation.
- Submitted by
- Michael Snyder
- Lab
- Michael Snyder, Stanford
- Grant
- U54HG004558
ELK1 (Homo sapiens)
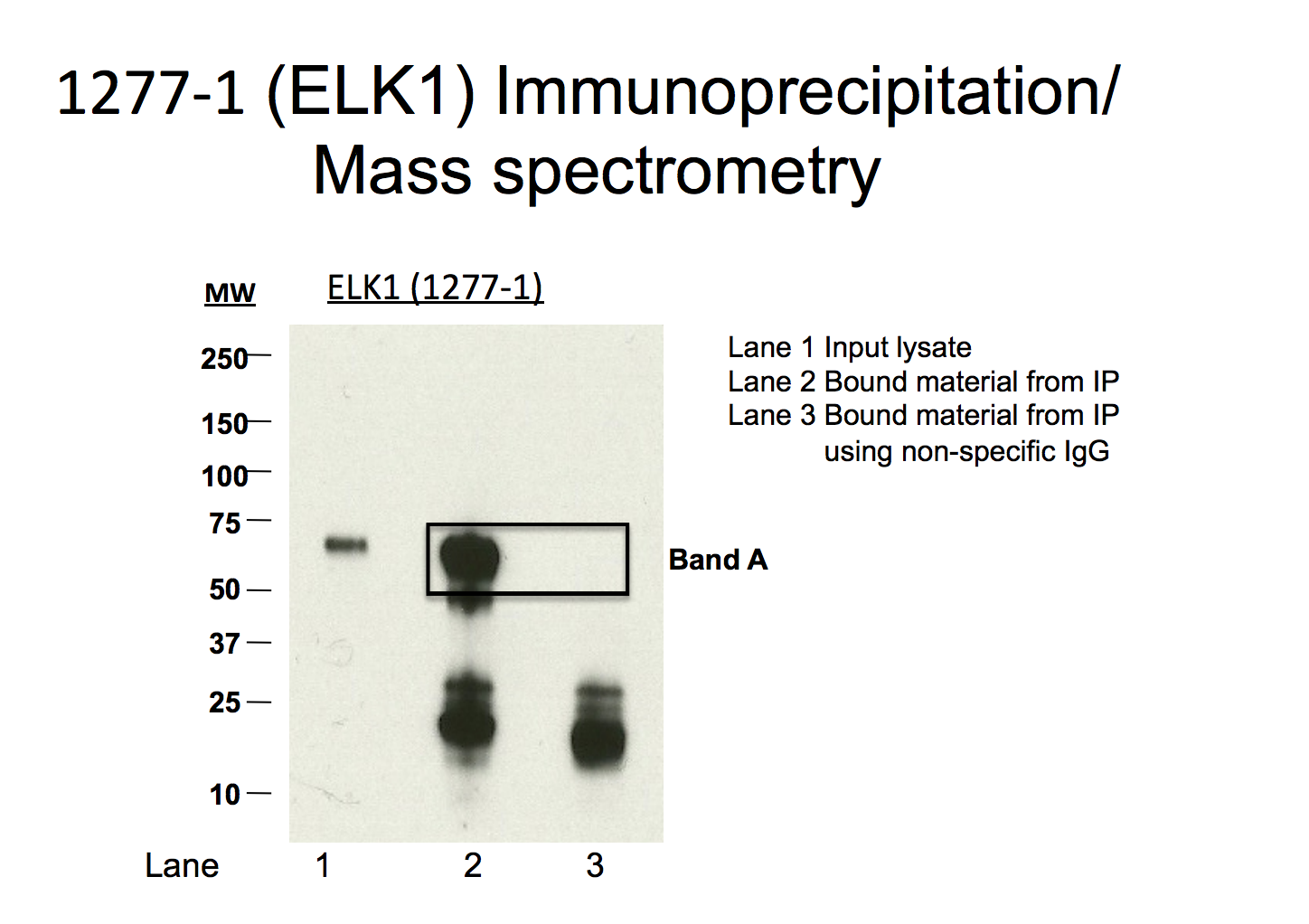
K562
compliant
- Caption
- Immunoprecipitation of ELK1 from K562 cells using 12777-1. Lane 1: input nuclear lysate. Lane 2: material immunoprecipitated with 1277-1. Lane 3: material immunoprecipitated using control IgG. Band A was excised from gel and subject to analysis by mass spectrometry.
- Submitted by
- Kathrina Onate
- Lab
- Michael Snyder, Stanford
- Grant
- U54HG004558
- Download
- IP_K562 Snyder AGB.png