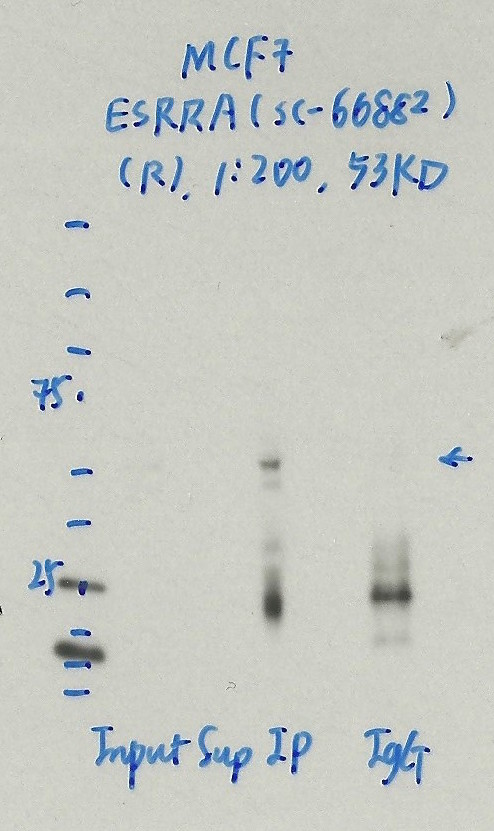ENCAB000AGE
Antibody against Homo sapiens ESRRA
Homo sapiens
MCF-7, HepG2, A549, K562
characterized to standards
Homo sapiens
any cell type or tissue
partially characterized
- Status
- released
- Source (vendor)
- Santa Cruz Biotech
- Product ID
- sc-66882
- Lot ID
- A0809
- Characterized targets
- ESRRA (Homo sapiens)
- Host
- rabbit
- Clonality
- polyclonal
- Purification
- other
- Isotype
- IgG
- Antigen description
- Epitope corresponding to amino acids 81-160 mapping near the N-terminus of ERRalpha of human origin
- External resources
Characterizations
ESRRA (Homo sapiens)
A549
compliant
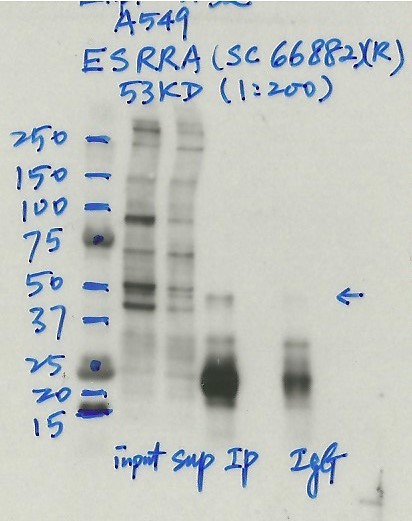
- Caption
- Immunoprecipitation was performed on nuclear extracts from the cell line: A549, using the antibody sc-66882. The blot shows western blot analysis of input, flowthrough, immunoprecipitate and mock immunoprecipitate using IgG.Molecular Weight: 53
- Submitted by
- Nathaniel Watson
- Lab
- Michael Snyder, Stanford
- Grant
- U54HG006996
- Download
- #1132 A549 ESRRA(sc66882).jpg
ESRRA (Homo sapiens)
MCF-7
compliant
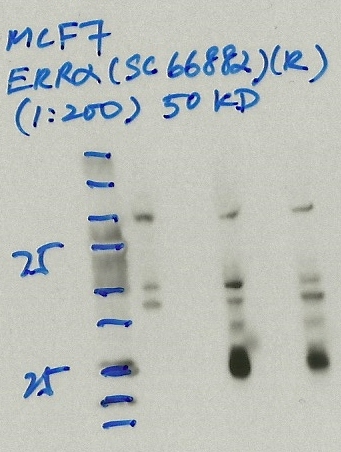
- Caption
- Immunoprecipitation was performed on nuclear extracts from the cell line: MCF-7, using the antibody sc-66882. The blot shows western blot analysis of input, flowthrough, immunoprecipitate and mock immunoprecipitate using IgG.Molecular Weight: 53
- Submitted by
- Nathaniel Watson
- Lab
- Michael Snyder, Stanford
- Grant
- U54HG006996
- Download
- 1128_03_ERRA_sc-66882.jpg
ESRRA (Homo sapiens)
MCF-7
not compliant
- Caption
- Immunoprecipitation was performed on nuclear extracts from the cell line: MCF-7, using the antibody sc-66882. The blot shows western blot analysis of input, flowthrough, immunoprecipitate and mock immunoprecipitate using IgG.
- Reviewer comment
- Same band in IgG lane.
- Submitted by
- Denis Salins
- Lab
- Michael Snyder, Stanford
- Grant
- U54HG006996
- Download
- 811 2_ ERRa.jpg
ESRRA (Homo sapiens)
Method: immunoprecipitation followed by mass spectrometry
compliant
- Caption
- IP followed by mass spectrometry. Briefly, protein was immunoprecipitated from K562 nuclear cell lysates using the antibody sc-66882, and the IP fraction was loaded on a 10% polyacrylamide gel (NuPAGEBis-Tris Gel) and separated with an Invitrogen NuPAGE electrophoresis system. The gel was stained by ColloidialCoomassie G-250 stain, gel fragments corresponding to the bands indicated were excised. Then proteins were trypsinized using the in-gel digestion method. Digested proteins were analyzed on an Orbitrap Elite mass spectrometer (Thermo Scientific) by the nanoLC-ESI-MS/MS technique. Peptides were identified by the SEQUEST algorithm and filtered with a high confidence threshold (Peptide false discovery rate < 1%, 2 unique peptides per protein minimum, mass error < 10 ppm).
- Submitter comment
- ESRRA acts as a site-specific transcription regulator and has been also shown to interact with estrogen and the transcripton factor TFIIB by direct protein-protein contact. http://www.genecards.org/cgi-bin/carddisp.pl?gene=ESRRA&keywords=esrra PUF60 is in association with FUBP1 regulates MYC transcription at the P2 promoter through the core-TFIIH basal transcription factor. Acts as a transcriptional repressor through the core-TFIIH basal transcription factor. http://www.genecards.org/cgi-bin/carddisp.pl?gene=PUF60&keywords=puf60. Both TFIIB and TFIIH are components of preinitiation complex (abbreviated PIC) which is a large complex of proteins that is necessary for the transcription of protein-coding genes in eukaryotes and archaea. The minimal PIC includes RNA polymerase II and six general transcription factors: TFIIA, TFIIB, TFIID, TFIIE, TFIIF, and TFIIH. https://en.wikipedia.org/wiki/Transcription_preinitiation_complex. PUF60 is 60KD, is in the ESRRA gel slice, but TFIIB (35KD) and TFIIH (ERCC3) (89KD) are not in the gel slice.
- Submitted by
- Nathaniel Watson
- Lab
- Michael Snyder, Stanford
- Grant
- U54HG006996
- Download
- ESRRA_sc-66882 final.pdf
ESRRA (Homo sapiens)
K562
compliant
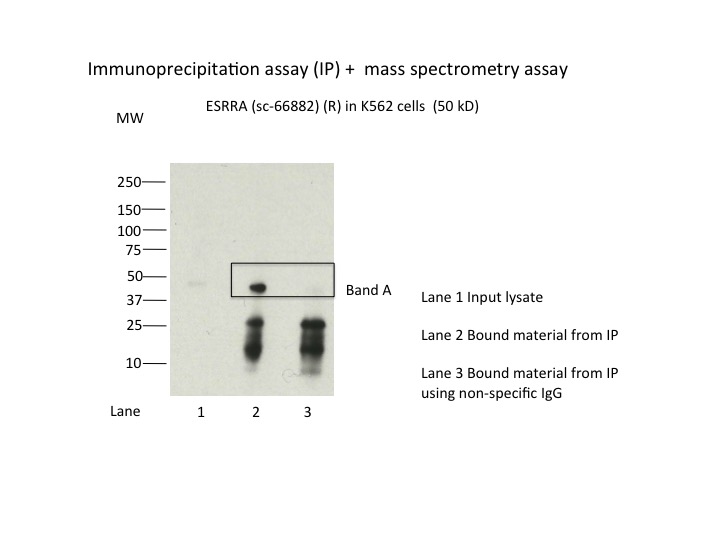
- Caption
- Immunoprecipitation was performed on nuclear extracts from the cell line K562 using the antibody sc-66882. Lane 1: input nuclear lysate. Lane 2: material immunoprecipitated with antibody. Lane 3: material immunoprecipitated using control IgG. Marked bands were excised from gel and subjected to analysis by mass spectrometry. Target molecular weight: 53.
- Submitted by
- Nathaniel Watson
- Lab
- Michael Snyder, Stanford
- Grant
- U54HG006996
- Download
- ESRRA_sc-66882.jpg
ESRRA (Homo sapiens)
Method: motif enrichment
not reviewed
- Caption
- Calculations were done by Pouya Kheradpour using a collection of known motifs available at http://www.broadinstitute.org/%7Epouyak/motif-disc/human/. Table 1 shows the fold-enrichments, enrichment p-values and fraction of peaks which contain the motif. The motif which produced the largest value for each criterion is shown in Table 1. Motifs were identified using a matching stringency corresponding to 4-6 (6-mer). Peaks identified by IDR (1% cutoff) were used in the analysis and +/-50bp from peak centers were considered. Enrichments are for a given motif vs. a background consisting of +/- 50bp from the centers of all DnaseI hypersensitive peaks. Repeat mask/simple repeats from UCSC and all gencode v7 exons (including non-protein coding genes) were excluded from the analysis. Comparison to shuffle motifs were used to correct for compositional bias. Enrichment is the corrected # of motifs in ChIP peaks/corrected # of motifs in DNaseI peaks. The current ENCODE standard calls for >4-fold enrichment and >10% motif representation for this criteria to be used for validation. The ERRA dataset presented here exceeds these thresholds and the antibody is considered validated.
- Submitted by
- Michael Snyder
- Lab
- Michael Snyder, Stanford
- Grant
- U54HG004558
- Download
- human_ERRA_validation_Snyder.pdf
ESRRA (Homo sapiens)
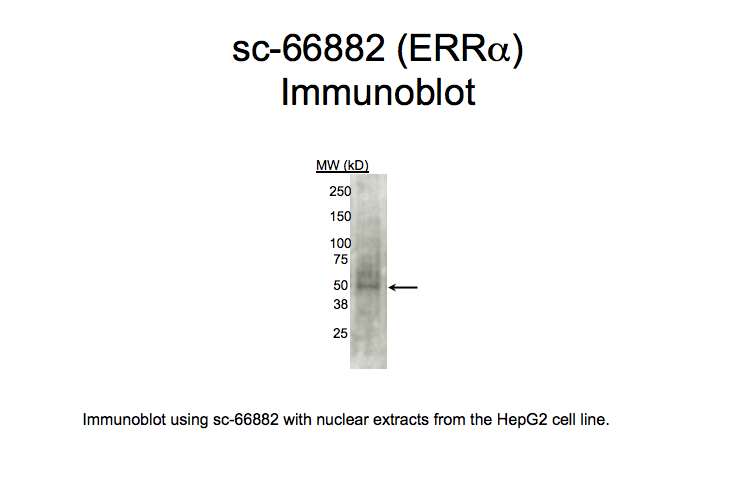
Method: immunoblot
not reviewed
- Caption
- A primary band with a mobility consistent with the size expected for ERRa (46kD; indicated by arrow) is identified by immunoblot with nuclear lysates from HepG2 cells. Therefore, sc-66882 meets this criterion for validation.
- Submitted by
- Michael Snyder
- Lab
- Michael Snyder, Stanford
- Grant
- U54HG004558
- Download
- human_ERRA_validation_Snyder.pdf
ESRRA (Homo sapiens)
HepG2
compliant
- Caption
- Immunoblot using sc-66882 with nuclear extracts from HepG2 cell line. Expected band size is 46 kD, indicated by arrow.
- Submitted by
- Kathrina Onate
- Lab
- Michael Snyder, Stanford
- Grant
- U54HG004558
- Download
- WB-HepG2 Snyder AGE.png